Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
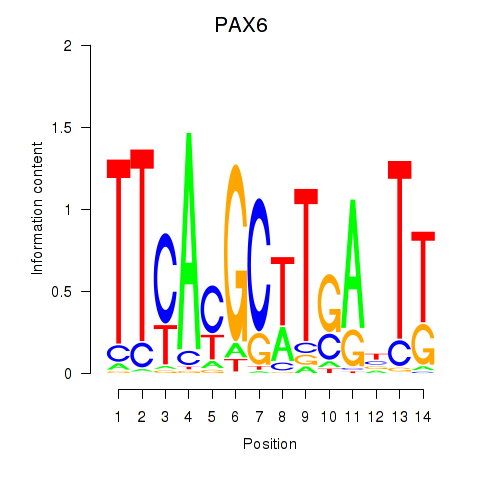
Results for PAX6
Z-value: 1.01
Transcription factors associated with PAX6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PAX6
|
ENSG00000007372.16 | PAX6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PAX6 | hg19_v2_chr11_-_31839488_31839515 | -0.26 | 5.4e-01 | Click! |
Activity profile of PAX6 motif
Sorted Z-values of PAX6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PAX6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_61420169 | 1.57 |
ENST00000425392.1 ENST00000336429.2 |
SERPINB7 |
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr11_-_87908600 | 1.41 |
ENST00000531138.1 ENST00000526372.1 ENST00000243662.6 |
RAB38 |
RAB38, member RAS oncogene family |
| chr18_+_61445007 | 1.34 |
ENST00000447428.1 ENST00000546027.1 |
SERPINB7 |
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr6_+_125474795 | 1.29 |
ENST00000304877.13 ENST00000534000.1 ENST00000368402.5 ENST00000368388.2 |
TPD52L1 |
tumor protein D52-like 1 |
| chr1_+_2005425 | 1.28 |
ENST00000461106.2 |
PRKCZ |
protein kinase C, zeta |
| chr6_+_125474992 | 1.23 |
ENST00000528193.1 |
TPD52L1 |
tumor protein D52-like 1 |
| chr6_+_125475335 | 1.22 |
ENST00000532429.1 ENST00000534199.1 |
TPD52L1 |
tumor protein D52-like 1 |
| chr2_-_161056762 | 1.19 |
ENST00000428609.2 ENST00000409967.2 |
ITGB6 |
integrin, beta 6 |
| chr6_+_125474939 | 1.19 |
ENST00000527711.1 |
TPD52L1 |
tumor protein D52-like 1 |
| chrX_-_106449656 | 1.18 |
ENST00000372466.4 ENST00000421752.1 ENST00000372461.3 |
NUP62CL |
nucleoporin 62kDa C-terminal like |
| chr2_-_161056802 | 1.15 |
ENST00000283249.2 ENST00000409872.1 |
ITGB6 |
integrin, beta 6 |
| chr1_-_201368653 | 1.07 |
ENST00000367313.3 |
LAD1 |
ladinin 1 |
| chr1_-_201368707 | 1.07 |
ENST00000391967.2 |
LAD1 |
ladinin 1 |
| chr4_+_77356248 | 1.05 |
ENST00000296043.6 |
SHROOM3 |
shroom family member 3 |
| chr9_+_93589734 | 0.98 |
ENST00000375746.1 |
SYK |
spleen tyrosine kinase |
| chr14_+_61995722 | 0.76 |
ENST00000556347.1 |
RP11-47I22.4 |
RP11-47I22.4 |
| chr12_+_47473369 | 0.73 |
ENST00000546455.1 |
PCED1B |
PC-esterase domain containing 1B |
| chr12_+_83080659 | 0.71 |
ENST00000321196.3 |
TMTC2 |
transmembrane and tetratricopeptide repeat containing 2 |
| chr1_+_77333117 | 0.71 |
ENST00000477717.1 |
ST6GALNAC5 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
| chr16_+_618837 | 0.71 |
ENST00000409439.2 |
PIGQ |
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr6_+_1080164 | 0.70 |
ENST00000314040.1 |
AL033381.1 |
Uncharacterized protein; cDNA FLJ34594 fis, clone KIDNE2009109 |
| chr5_+_140254884 | 0.62 |
ENST00000398631.2 |
PCDHA12 |
protocadherin alpha 12 |
| chr11_-_58343319 | 0.59 |
ENST00000395074.2 |
LPXN |
leupaxin |
| chr12_-_85306562 | 0.59 |
ENST00000551612.1 ENST00000450363.3 ENST00000552192.1 |
SLC6A15 |
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr3_-_124839648 | 0.56 |
ENST00000430155.2 |
SLC12A8 |
solute carrier family 12, member 8 |
| chr8_+_21916680 | 0.51 |
ENST00000358242.3 ENST00000415253.1 |
DMTN |
dematin actin binding protein |
| chr11_+_128563652 | 0.50 |
ENST00000527786.2 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr1_-_185597619 | 0.49 |
ENST00000608417.1 ENST00000436955.1 |
GS1-204I12.1 |
GS1-204I12.1 |
| chr8_+_21916710 | 0.49 |
ENST00000523266.1 ENST00000519907.1 |
DMTN |
dematin actin binding protein |
| chr10_-_98031265 | 0.47 |
ENST00000224337.5 ENST00000371176.2 |
BLNK |
B-cell linker |
| chr12_+_113354341 | 0.46 |
ENST00000553152.1 |
OAS1 |
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr10_-_118765081 | 0.42 |
ENST00000392903.2 ENST00000355371.4 |
KIAA1598 |
KIAA1598 |
| chr12_-_85306594 | 0.42 |
ENST00000266682.5 |
SLC6A15 |
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr2_+_29320571 | 0.42 |
ENST00000401605.1 ENST00000401617.2 |
CLIP4 |
CAP-GLY domain containing linker protein family, member 4 |
| chr8_-_95487272 | 0.39 |
ENST00000297592.5 |
RAD54B |
RAD54 homolog B (S. cerevisiae) |
| chr11_+_65554493 | 0.39 |
ENST00000335987.3 |
OVOL1 |
ovo-like zinc finger 1 |
| chr20_-_44144249 | 0.39 |
ENST00000217428.6 |
SPINT3 |
serine peptidase inhibitor, Kunitz type, 3 |
| chr19_+_49891475 | 0.37 |
ENST00000447857.3 |
CCDC155 |
coiled-coil domain containing 155 |
| chr16_+_84682108 | 0.35 |
ENST00000564996.1 ENST00000258157.5 ENST00000567410.1 |
KLHL36 |
kelch-like family member 36 |
| chr11_-_117747434 | 0.35 |
ENST00000529335.2 ENST00000530956.1 ENST00000260282.4 |
FXYD6 |
FXYD domain containing ion transport regulator 6 |
| chr10_-_118764862 | 0.35 |
ENST00000260777.10 |
KIAA1598 |
KIAA1598 |
| chr11_-_117748138 | 0.34 |
ENST00000527717.1 |
FXYD6 |
FXYD domain containing ion transport regulator 6 |
| chr12_-_71031220 | 0.34 |
ENST00000334414.6 |
PTPRB |
protein tyrosine phosphatase, receptor type, B |
| chr11_-_117747607 | 0.33 |
ENST00000540359.1 ENST00000539526.1 |
FXYD6 |
FXYD domain containing ion transport regulator 6 |
| chr6_-_134373732 | 0.33 |
ENST00000275230.5 |
SLC2A12 |
solute carrier family 2 (facilitated glucose transporter), member 12 |
| chr20_-_62178857 | 0.32 |
ENST00000217188.1 |
SRMS |
src-related kinase lacking C-terminal regulatory tyrosine and N-terminal myristylation sites |
| chr17_-_39023462 | 0.32 |
ENST00000251643.4 |
KRT12 |
keratin 12 |
| chr2_+_196521458 | 0.32 |
ENST00000409086.3 |
SLC39A10 |
solute carrier family 39 (zinc transporter), member 10 |
| chr13_+_96085847 | 0.30 |
ENST00000376873.3 |
CLDN10 |
claudin 10 |
| chr6_-_10412600 | 0.29 |
ENST00000379608.3 |
TFAP2A |
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
| chr2_+_196521845 | 0.29 |
ENST00000359634.5 ENST00000412905.1 |
SLC39A10 |
solute carrier family 39 (zinc transporter), member 10 |
| chr3_-_196910721 | 0.28 |
ENST00000443183.1 |
DLG1 |
discs, large homolog 1 (Drosophila) |
| chrX_+_37545012 | 0.28 |
ENST00000378616.3 |
XK |
X-linked Kx blood group (McLeod syndrome) |
| chr1_+_73771844 | 0.28 |
ENST00000440762.1 ENST00000444827.1 ENST00000415686.1 ENST00000411903.1 |
RP4-598G3.1 |
RP4-598G3.1 |
| chr17_-_9694614 | 0.28 |
ENST00000330255.5 ENST00000571134.1 |
DHRS7C |
dehydrogenase/reductase (SDR family) member 7C |
| chr1_+_161677034 | 0.27 |
ENST00000349527.4 ENST00000309691.6 ENST00000294796.4 ENST00000367953.3 ENST00000367950.1 |
FCRLA |
Fc receptor-like A |
| chr19_+_41222998 | 0.27 |
ENST00000263370.2 |
ITPKC |
inositol-trisphosphate 3-kinase C |
| chr3_+_140981456 | 0.27 |
ENST00000504264.1 |
ACPL2 |
acid phosphatase-like 2 |
| chr8_-_21771182 | 0.27 |
ENST00000523932.1 ENST00000544659.1 |
DOK2 |
docking protein 2, 56kDa |
| chr8_-_21771214 | 0.27 |
ENST00000276420.4 |
DOK2 |
docking protein 2, 56kDa |
| chr2_+_192110199 | 0.27 |
ENST00000304164.4 |
MYO1B |
myosin IB |
| chr10_+_12110963 | 0.27 |
ENST00000263035.4 ENST00000437298.1 |
DHTKD1 |
dehydrogenase E1 and transketolase domain containing 1 |
| chr15_+_68924327 | 0.26 |
ENST00000543950.1 |
CORO2B |
coronin, actin binding protein, 2B |
| chr4_+_76995855 | 0.26 |
ENST00000355810.4 ENST00000349321.3 |
ART3 |
ADP-ribosyltransferase 3 |
| chr2_+_192109911 | 0.25 |
ENST00000418908.1 ENST00000339514.4 ENST00000392318.3 |
MYO1B |
myosin IB |
| chrX_-_130423200 | 0.25 |
ENST00000361420.3 |
IGSF1 |
immunoglobulin superfamily, member 1 |
| chrX_-_130423386 | 0.24 |
ENST00000370903.3 |
IGSF1 |
immunoglobulin superfamily, member 1 |
| chr4_-_88141755 | 0.24 |
ENST00000273963.5 |
KLHL8 |
kelch-like family member 8 |
| chr12_-_15103621 | 0.24 |
ENST00000536592.1 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
| chr3_-_111852128 | 0.23 |
ENST00000308910.4 |
GCSAM |
germinal center-associated, signaling and motility |
| chr3_-_122512619 | 0.23 |
ENST00000383659.1 ENST00000306103.2 |
HSPBAP1 |
HSPB (heat shock 27kDa) associated protein 1 |
| chr2_+_232135245 | 0.23 |
ENST00000446447.1 |
ARMC9 |
armadillo repeat containing 9 |
| chr17_+_57297807 | 0.22 |
ENST00000284116.4 ENST00000581140.1 ENST00000581276.1 |
GDPD1 |
glycerophosphodiester phosphodiesterase domain containing 1 |
| chr19_-_41222775 | 0.22 |
ENST00000324464.3 ENST00000450541.1 ENST00000594720.1 |
ADCK4 |
aarF domain containing kinase 4 |
| chr15_+_52155001 | 0.22 |
ENST00000544199.1 |
TMOD3 |
tropomodulin 3 (ubiquitous) |
| chr2_-_136743169 | 0.22 |
ENST00000264161.4 |
DARS |
aspartyl-tRNA synthetase |
| chr1_+_180897269 | 0.21 |
ENST00000367587.1 |
KIAA1614 |
KIAA1614 |
| chr19_-_36019123 | 0.21 |
ENST00000588674.1 ENST00000452271.2 ENST00000518157.1 |
SBSN |
suprabasin |
| chr16_+_31128978 | 0.21 |
ENST00000448516.2 ENST00000219797.4 |
KAT8 |
K(lysine) acetyltransferase 8 |
| chr7_+_76109827 | 0.20 |
ENST00000446820.2 |
DTX2 |
deltex homolog 2 (Drosophila) |
| chr16_+_84801852 | 0.20 |
ENST00000569925.1 ENST00000567526.1 |
USP10 |
ubiquitin specific peptidase 10 |
| chrX_-_138914394 | 0.20 |
ENST00000327569.3 ENST00000361648.2 ENST00000370543.1 ENST00000359686.2 |
ATP11C |
ATPase, class VI, type 11C |
| chr4_-_88141615 | 0.20 |
ENST00000545252.1 ENST00000425278.2 ENST00000498875.2 |
KLHL8 |
kelch-like family member 8 |
| chr14_+_24867992 | 0.20 |
ENST00000382554.3 |
NYNRIN |
NYN domain and retroviral integrase containing |
| chr11_+_118175132 | 0.19 |
ENST00000361763.4 |
CD3E |
CD3e molecule, epsilon (CD3-TCR complex) |
| chr4_-_174320687 | 0.19 |
ENST00000296506.3 |
SCRG1 |
stimulator of chondrogenesis 1 |
| chr6_-_42162654 | 0.19 |
ENST00000230361.3 |
GUCA1B |
guanylate cyclase activator 1B (retina) |
| chr19_-_41903161 | 0.18 |
ENST00000602129.1 ENST00000593771.1 ENST00000596905.1 ENST00000221233.4 |
EXOSC5 |
exosome component 5 |
| chr4_+_40751914 | 0.18 |
ENST00000381782.2 ENST00000316607.5 |
NSUN7 |
NOP2/Sun domain family, member 7 |
| chr7_-_80551671 | 0.18 |
ENST00000419255.2 ENST00000544525.1 |
SEMA3C |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr15_-_85201779 | 0.18 |
ENST00000360476.3 ENST00000394588.3 |
NMB |
neuromedin B |
| chr14_+_22782867 | 0.18 |
ENST00000390467.3 |
TRAV40 |
T cell receptor alpha variable 40 |
| chr3_+_15045419 | 0.18 |
ENST00000406272.2 |
NR2C2 |
nuclear receptor subfamily 2, group C, member 2 |
| chr16_-_71264558 | 0.17 |
ENST00000448089.2 ENST00000393550.2 ENST00000448691.1 ENST00000393567.2 ENST00000321489.5 ENST00000539973.1 ENST00000288168.10 ENST00000545267.1 ENST00000541601.1 ENST00000538248.1 |
HYDIN |
HYDIN, axonemal central pair apparatus protein |
| chr2_+_145780725 | 0.17 |
ENST00000451478.1 |
TEX41 |
testis expressed 41 (non-protein coding) |
| chr12_-_70093235 | 0.17 |
ENST00000266661.4 |
BEST3 |
bestrophin 3 |
| chr2_+_238877424 | 0.17 |
ENST00000434655.1 |
UBE2F |
ubiquitin-conjugating enzyme E2F (putative) |
| chrX_+_106449862 | 0.17 |
ENST00000372453.3 ENST00000535523.1 |
PIH1D3 |
PIH1 domain containing 3 |
| chr4_+_129349188 | 0.17 |
ENST00000511497.1 |
RP11-420A23.1 |
RP11-420A23.1 |
| chr11_-_18813110 | 0.17 |
ENST00000396168.1 |
PTPN5 |
protein tyrosine phosphatase, non-receptor type 5 (striatum-enriched) |
| chr17_-_2415169 | 0.16 |
ENST00000263092.6 ENST00000538844.1 ENST00000576976.1 |
METTL16 |
methyltransferase like 16 |
| chr5_+_40909354 | 0.16 |
ENST00000313164.9 |
C7 |
complement component 7 |
| chr1_+_207925391 | 0.16 |
ENST00000358170.2 ENST00000354848.1 ENST00000322918.5 ENST00000367042.1 ENST00000367041.1 ENST00000357714.1 ENST00000322875.4 ENST00000367047.1 ENST00000441839.2 ENST00000361067.1 ENST00000360212.2 ENST00000480003.1 |
CD46 |
CD46 molecule, complement regulatory protein |
| chr13_+_25875662 | 0.16 |
ENST00000381736.3 ENST00000463407.1 ENST00000381718.3 |
NUPL1 |
nucleoporin like 1 |
| chr12_+_6949964 | 0.15 |
ENST00000541978.1 ENST00000435982.2 |
GNB3 |
guanine nucleotide binding protein (G protein), beta polypeptide 3 |
| chr8_-_95487331 | 0.15 |
ENST00000336148.5 |
RAD54B |
RAD54 homolog B (S. cerevisiae) |
| chr16_-_1401799 | 0.15 |
ENST00000007390.2 |
TSR3 |
TSR3, 20S rRNA accumulation, homolog (S. cerevisiae) |
| chr5_+_85913721 | 0.15 |
ENST00000247655.3 ENST00000509578.1 ENST00000515763.1 |
COX7C |
cytochrome c oxidase subunit VIIc |
| chr16_-_3767551 | 0.15 |
ENST00000246957.5 |
TRAP1 |
TNF receptor-associated protein 1 |
| chr21_+_39628852 | 0.15 |
ENST00000398938.2 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr18_-_45663666 | 0.15 |
ENST00000535628.2 |
ZBTB7C |
zinc finger and BTB domain containing 7C |
| chr1_-_87380002 | 0.15 |
ENST00000331835.5 |
SEP15 |
Homo sapiens 15 kDa selenoprotein (SEP15), transcript variant 2, mRNA. |
| chr2_-_29093132 | 0.15 |
ENST00000306108.5 |
TRMT61B |
tRNA methyltransferase 61 homolog B (S. cerevisiae) |
| chr18_+_13611431 | 0.15 |
ENST00000587757.1 |
LDLRAD4 |
low density lipoprotein receptor class A domain containing 4 |
| chrX_-_124097620 | 0.14 |
ENST00000371130.3 ENST00000422452.2 |
TENM1 |
teneurin transmembrane protein 1 |
| chr4_-_87281224 | 0.14 |
ENST00000395169.3 ENST00000395161.2 |
MAPK10 |
mitogen-activated protein kinase 10 |
| chr1_-_85725316 | 0.14 |
ENST00000344356.5 ENST00000471115.1 |
C1orf52 |
chromosome 1 open reading frame 52 |
| chr17_-_77023723 | 0.14 |
ENST00000577521.1 |
C1QTNF1-AS1 |
C1QTNF1 antisense RNA 1 |
| chr1_+_147374915 | 0.14 |
ENST00000240986.4 |
GJA8 |
gap junction protein, alpha 8, 50kDa |
| chr11_-_18813353 | 0.13 |
ENST00000358540.2 ENST00000396171.4 ENST00000396167.2 |
PTPN5 |
protein tyrosine phosphatase, non-receptor type 5 (striatum-enriched) |
| chr1_-_55089191 | 0.13 |
ENST00000302250.2 ENST00000371304.2 |
FAM151A |
family with sequence similarity 151, member A |
| chr16_-_67450325 | 0.13 |
ENST00000348579.2 |
ZDHHC1 |
zinc finger, DHHC-type containing 1 |
| chr1_-_156722015 | 0.13 |
ENST00000368209.5 |
HDGF |
hepatoma-derived growth factor |
| chr17_-_77023683 | 0.13 |
ENST00000581579.1 |
C1QTNF1-AS1 |
C1QTNF1 antisense RNA 1 |
| chr5_-_141338627 | 0.12 |
ENST00000231484.3 |
PCDH12 |
protocadherin 12 |
| chr2_-_70520539 | 0.12 |
ENST00000482975.2 ENST00000438261.1 |
SNRPG |
small nuclear ribonucleoprotein polypeptide G |
| chr4_+_187148556 | 0.12 |
ENST00000264690.6 ENST00000446598.2 ENST00000414291.1 ENST00000513864.1 |
KLKB1 |
kallikrein B, plasma (Fletcher factor) 1 |
| chr12_+_111284805 | 0.12 |
ENST00000552694.1 |
CCDC63 |
coiled-coil domain containing 63 |
| chr5_-_55529115 | 0.12 |
ENST00000513241.2 ENST00000341048.4 |
ANKRD55 |
ankyrin repeat domain 55 |
| chr11_-_117747327 | 0.12 |
ENST00000584230.1 ENST00000527429.1 ENST00000584394.1 ENST00000532984.1 |
FXYD6 FXYD6-FXYD2 |
FXYD domain containing ion transport regulator 6 FXYD6-FXYD2 readthrough |
| chr22_+_26138108 | 0.12 |
ENST00000536101.1 ENST00000335473.7 ENST00000407587.2 |
MYO18B |
myosin XVIIIB |
| chr11_+_28129795 | 0.11 |
ENST00000406787.3 ENST00000342303.5 ENST00000403099.1 ENST00000407364.3 |
METTL15 |
methyltransferase like 15 |
| chr10_-_75226166 | 0.11 |
ENST00000544628.1 |
PPP3CB |
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr19_+_3708376 | 0.11 |
ENST00000539908.2 |
TJP3 |
tight junction protein 3 |
| chr11_-_11374904 | 0.11 |
ENST00000528848.2 |
CSNK2A3 |
casein kinase 2, alpha 3 polypeptide |
| chr19_+_35939154 | 0.11 |
ENST00000599180.2 |
FFAR2 |
free fatty acid receptor 2 |
| chr12_+_56915713 | 0.11 |
ENST00000262031.5 ENST00000552247.2 |
RBMS2 |
RNA binding motif, single stranded interacting protein 2 |
| chr12_+_111284764 | 0.11 |
ENST00000545036.1 ENST00000308208.5 |
CCDC63 |
coiled-coil domain containing 63 |
| chr3_+_51663407 | 0.11 |
ENST00000432863.1 ENST00000296477.3 |
RAD54L2 |
RAD54-like 2 (S. cerevisiae) |
| chr12_+_56915776 | 0.11 |
ENST00000550726.1 ENST00000542360.1 |
RBMS2 |
RNA binding motif, single stranded interacting protein 2 |
| chr16_-_3767506 | 0.11 |
ENST00000538171.1 |
TRAP1 |
TNF receptor-associated protein 1 |
| chr13_+_25875785 | 0.11 |
ENST00000381747.3 |
NUPL1 |
nucleoporin like 1 |
| chr9_+_87283430 | 0.10 |
ENST00000376214.1 ENST00000376213.1 |
NTRK2 |
neurotrophic tyrosine kinase, receptor, type 2 |
| chr6_-_35480640 | 0.10 |
ENST00000428978.1 ENST00000322263.4 |
TULP1 |
tubby like protein 1 |
| chr11_-_3078838 | 0.10 |
ENST00000397111.5 |
CARS |
cysteinyl-tRNA synthetase |
| chr2_-_86116093 | 0.10 |
ENST00000377332.3 |
ST3GAL5 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr6_-_137366163 | 0.10 |
ENST00000367748.1 |
IL20RA |
interleukin 20 receptor, alpha |
| chr1_+_167298281 | 0.10 |
ENST00000367862.5 |
POU2F1 |
POU class 2 homeobox 1 |
| chr17_-_39216344 | 0.10 |
ENST00000391418.2 |
KRTAP2-3 |
keratin associated protein 2-3 |
| chr2_-_99870744 | 0.10 |
ENST00000409238.1 ENST00000423800.1 |
LYG2 |
lysozyme G-like 2 |
| chr7_-_87342564 | 0.10 |
ENST00000265724.3 ENST00000416177.1 |
ABCB1 |
ATP-binding cassette, sub-family B (MDR/TAP), member 1 |
| chr9_-_128246769 | 0.10 |
ENST00000444226.1 |
MAPKAP1 |
mitogen-activated protein kinase associated protein 1 |
| chr10_+_97471508 | 0.09 |
ENST00000453258.2 |
ENTPD1 |
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr6_-_33041378 | 0.09 |
ENST00000428995.1 |
HLA-DPA1 |
major histocompatibility complex, class II, DP alpha 1 |
| chr2_-_163008903 | 0.09 |
ENST00000418842.2 ENST00000375497.3 |
GCG |
glucagon |
| chr4_+_113970772 | 0.09 |
ENST00000504454.1 ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2 |
ankyrin 2, neuronal |
| chr12_-_69080590 | 0.09 |
ENST00000433116.2 ENST00000500695.2 |
RP11-637A17.2 |
RP11-637A17.2 |
| chr19_-_50370509 | 0.09 |
ENST00000596014.1 |
PNKP |
polynucleotide kinase 3'-phosphatase |
| chr4_+_159236462 | 0.09 |
ENST00000460056.2 |
RXFP1 |
relaxin/insulin-like family peptide receptor 1 |
| chr4_+_70861647 | 0.09 |
ENST00000246895.4 ENST00000381060.2 |
STATH |
statherin |
| chr11_+_4788500 | 0.09 |
ENST00000380390.1 |
MMP26 |
matrix metallopeptidase 26 |
| chr14_+_24407940 | 0.09 |
ENST00000354854.1 |
DHRS4-AS1 |
DHRS4-AS1 |
| chr12_+_69080734 | 0.09 |
ENST00000378905.2 |
NUP107 |
nucleoporin 107kDa |
| chr9_-_128412696 | 0.09 |
ENST00000420643.1 |
MAPKAP1 |
mitogen-activated protein kinase associated protein 1 |
| chr21_-_18985230 | 0.09 |
ENST00000457956.1 ENST00000348354.6 |
BTG3 |
BTG family, member 3 |
| chr12_-_10007448 | 0.09 |
ENST00000538152.1 |
CLEC2B |
C-type lectin domain family 2, member B |
| chr1_+_15668240 | 0.09 |
ENST00000444385.1 |
FHAD1 |
forkhead-associated (FHA) phosphopeptide binding domain 1 |
| chr6_+_42896865 | 0.08 |
ENST00000372836.4 ENST00000394142.3 |
CNPY3 |
canopy FGF signaling regulator 3 |
| chr16_+_77233294 | 0.08 |
ENST00000378644.4 |
SYCE1L |
synaptonemal complex central element protein 1-like |
| chr7_-_43909090 | 0.08 |
ENST00000317534.5 |
MRPS24 |
mitochondrial ribosomal protein S24 |
| chr8_+_121457642 | 0.08 |
ENST00000305949.1 |
MTBP |
Mdm2, transformed 3T3 cell double minute 2, p53 binding protein (mouse) binding protein, 104kDa |
| chr4_-_68749699 | 0.08 |
ENST00000545541.1 |
TMPRSS11D |
transmembrane protease, serine 11D |
| chr8_-_121457608 | 0.08 |
ENST00000306185.3 |
MRPL13 |
mitochondrial ribosomal protein L13 |
| chr4_-_68749745 | 0.08 |
ENST00000283916.6 |
TMPRSS11D |
transmembrane protease, serine 11D |
| chr3_-_142297668 | 0.08 |
ENST00000350721.4 ENST00000383101.3 |
ATR |
ataxia telangiectasia and Rad3 related |
| chr11_+_33563821 | 0.08 |
ENST00000321505.4 ENST00000265654.5 ENST00000389726.3 |
KIAA1549L |
KIAA1549-like |
| chr6_-_43027105 | 0.08 |
ENST00000230413.5 ENST00000487429.1 ENST00000489623.1 ENST00000468957.1 |
MRPL2 |
mitochondrial ribosomal protein L2 |
| chr2_+_168675182 | 0.07 |
ENST00000305861.1 |
B3GALT1 |
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr20_+_48807351 | 0.07 |
ENST00000303004.3 |
CEBPB |
CCAAT/enhancer binding protein (C/EBP), beta |
| chr22_+_39745930 | 0.07 |
ENST00000318801.4 ENST00000216155.7 ENST00000406293.3 ENST00000328933.5 |
SYNGR1 |
synaptogyrin 1 |
| chr2_+_145780767 | 0.07 |
ENST00000599358.1 ENST00000596278.1 ENST00000596747.1 ENST00000608652.1 ENST00000609705.1 ENST00000608432.1 ENST00000596970.1 ENST00000602041.1 ENST00000601578.1 ENST00000596034.1 ENST00000414195.2 ENST00000594837.1 |
TEX41 |
testis expressed 41 (non-protein coding) |
| chr8_-_68658578 | 0.07 |
ENST00000518549.1 ENST00000297770.4 ENST00000297769.4 |
CPA6 |
carboxypeptidase A6 |
| chr14_+_67291158 | 0.07 |
ENST00000555456.1 |
GPHN |
gephyrin |
| chr6_-_41039567 | 0.07 |
ENST00000468811.1 |
OARD1 |
O-acyl-ADP-ribose deacylase 1 |
| chr8_+_67104323 | 0.07 |
ENST00000518412.1 ENST00000518035.1 ENST00000517725.1 |
LINC00967 |
long intergenic non-protein coding RNA 967 |
| chr1_-_184006829 | 0.07 |
ENST00000361927.4 |
COLGALT2 |
collagen beta(1-O)galactosyltransferase 2 |
| chr12_-_47473425 | 0.07 |
ENST00000550413.1 |
AMIGO2 |
adhesion molecule with Ig-like domain 2 |
| chr6_-_137366096 | 0.07 |
ENST00000316649.5 ENST00000367746.3 |
IL20RA |
interleukin 20 receptor, alpha |
| chr4_-_65275162 | 0.07 |
ENST00000381210.3 ENST00000507440.1 |
TECRL |
trans-2,3-enoyl-CoA reductase-like |
| chr7_-_76255444 | 0.07 |
ENST00000454397.1 |
POMZP3 |
POM121 and ZP3 fusion |
| chr17_+_58499844 | 0.06 |
ENST00000269127.4 |
C17orf64 |
chromosome 17 open reading frame 64 |
| chr8_+_142402089 | 0.06 |
ENST00000521578.1 ENST00000520105.1 ENST00000523147.1 |
PTP4A3 |
protein tyrosine phosphatase type IVA, member 3 |
| chrX_+_144908928 | 0.06 |
ENST00000408967.2 |
TMEM257 |
transmembrane protein 257 |
| chr20_-_1472029 | 0.06 |
ENST00000359801.3 |
SIRPB2 |
signal-regulatory protein beta 2 |
| chr1_-_110933611 | 0.06 |
ENST00000472422.2 ENST00000437429.2 |
SLC16A4 |
solute carrier family 16, member 4 |
| chr5_-_98262240 | 0.06 |
ENST00000284049.3 |
CHD1 |
chromodomain helicase DNA binding protein 1 |
| chr7_+_112120908 | 0.06 |
ENST00000439068.2 ENST00000312849.4 ENST00000429049.1 |
LSMEM1 |
leucine-rich single-pass membrane protein 1 |
| chr4_-_10686373 | 0.06 |
ENST00000442825.2 |
CLNK |
cytokine-dependent hematopoietic cell linker |
| chr11_+_111789580 | 0.06 |
ENST00000278601.5 |
C11orf52 |
chromosome 11 open reading frame 52 |
| chr3_+_171561127 | 0.06 |
ENST00000334567.5 ENST00000450693.1 |
TMEM212 |
transmembrane protein 212 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.9 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.7 | 0.7 | GO:0050801 | ion homeostasis(GO:0050801) cation homeostasis(GO:0055080) inorganic ion homeostasis(GO:0098771) |
| 0.2 | 1.0 | GO:0045401 | response to molecule of fungal origin(GO:0002238) positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) cellular response to molecule of fungal origin(GO:0071226) |
| 0.2 | 2.3 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.2 | 1.4 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.2 | 1.3 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.2 | 1.0 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.1 | 0.4 | GO:0044771 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.1 | 1.0 | GO:0015820 | leucine transport(GO:0015820) proline transmembrane transport(GO:0035524) |
| 0.1 | 0.3 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.1 | 0.6 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.1 | 0.3 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.1 | 0.2 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.1 | 0.3 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.1 | 0.9 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.1 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.6 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.2 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.2 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.1 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.0 | 0.3 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.2 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.3 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.1 | GO:0006423 | cysteinyl-tRNA aminoacylation(GO:0006423) |
| 0.0 | 0.2 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.8 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.2 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.0 | 0.7 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.2 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 4.9 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 0.2 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.1 | GO:1904882 | telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.0 | 0.1 | GO:0042040 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.0 | 0.1 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.1 | GO:0070982 | L-asparagine biosynthetic process(GO:0070981) L-asparagine metabolic process(GO:0070982) |
| 0.0 | 0.4 | GO:0090220 | chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) |
| 0.0 | 0.1 | GO:0099538 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.0 | 0.5 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.1 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.2 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.1 | GO:0098502 | DNA dephosphorylation(GO:0098502) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.5 | GO:0006312 | mitotic recombination(GO:0006312) |
| 0.0 | 0.1 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.0 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.2 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 0.0 | 0.1 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.0 | 0.2 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 1.1 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.1 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.1 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 | 0.4 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.0 | 0.1 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 0.1 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.2 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.3 | 1.4 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.1 | 1.0 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 1.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 0.2 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.1 | 1.2 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 0.7 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 1.0 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.2 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.4 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.1 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.0 | 0.3 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.3 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 2.0 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.2 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.4 | GO:0035651 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.3 | 1.0 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.1 | 0.5 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.7 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 0.5 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 1.3 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.1 | 1.0 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.1 | 0.7 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 1.0 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.2 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 0.6 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.5 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.2 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 1.2 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.3 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 0.1 | GO:0004817 | cysteine-tRNA ligase activity(GO:0004817) |
| 0.0 | 0.3 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.1 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 0.2 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.0 | 0.2 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.2 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.0 | 3.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0061598 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.3 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 2.4 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.1 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.0 | 0.2 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.0 | 0.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.2 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.2 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.1 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.2 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.3 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.3 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.5 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 1.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 4.9 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.5 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 1.0 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.5 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.8 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 2.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.3 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |