Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
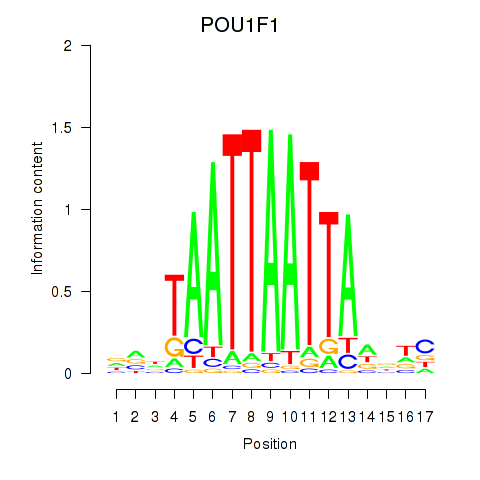
Results for POU1F1
Z-value: 0.73
Transcription factors associated with POU1F1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU1F1
|
ENSG00000064835.6 | POU1F1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU1F1 | hg19_v2_chr3_-_87325612_87325654 | -0.32 | 4.4e-01 | Click! |
Activity profile of POU1F1 motif
Sorted Z-values of POU1F1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of POU1F1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_119867159 | 0.74 |
ENST00000505123.1 |
PRR16 |
proline rich 16 |
| chr5_+_95066823 | 0.73 |
ENST00000506817.1 ENST00000379982.3 |
RHOBTB3 |
Rho-related BTB domain containing 3 |
| chr12_-_15038779 | 0.68 |
ENST00000228938.5 ENST00000539261.1 |
MGP |
matrix Gla protein |
| chr11_+_5710919 | 0.56 |
ENST00000379965.3 ENST00000425490.1 |
TRIM22 |
tripartite motif containing 22 |
| chr11_-_63376013 | 0.55 |
ENST00000540943.1 |
PLA2G16 |
phospholipase A2, group XVI |
| chr19_+_50016610 | 0.53 |
ENST00000596975.1 |
FCGRT |
Fc fragment of IgG, receptor, transporter, alpha |
| chr19_-_58864848 | 0.49 |
ENST00000263100.3 |
A1BG |
alpha-1-B glycoprotein |
| chr8_+_70404996 | 0.49 |
ENST00000402687.4 ENST00000419716.3 |
SULF1 |
sulfatase 1 |
| chr19_+_50016411 | 0.42 |
ENST00000426395.3 ENST00000600273.1 ENST00000599988.1 |
FCGRT |
Fc fragment of IgG, receptor, transporter, alpha |
| chr1_+_221051699 | 0.41 |
ENST00000366903.6 |
HLX |
H2.0-like homeobox |
| chr17_+_1674982 | 0.41 |
ENST00000572048.1 ENST00000573763.1 |
SERPINF1 |
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr2_+_152214098 | 0.39 |
ENST00000243347.3 |
TNFAIP6 |
tumor necrosis factor, alpha-induced protein 6 |
| chr2_-_190044480 | 0.37 |
ENST00000374866.3 |
COL5A2 |
collagen, type V, alpha 2 |
| chr5_+_156696362 | 0.36 |
ENST00000377576.3 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
| chr7_+_129932974 | 0.33 |
ENST00000445470.2 ENST00000222482.4 ENST00000492072.1 ENST00000473956.1 ENST00000493259.1 ENST00000486598.1 |
CPA4 |
carboxypeptidase A4 |
| chr6_-_52859046 | 0.33 |
ENST00000457564.1 ENST00000541324.1 ENST00000370960.1 |
GSTA4 |
glutathione S-transferase alpha 4 |
| chr3_-_191000172 | 0.31 |
ENST00000427544.2 |
UTS2B |
urotensin 2B |
| chr7_-_45960850 | 0.31 |
ENST00000381083.4 ENST00000381086.5 ENST00000275521.6 |
IGFBP3 |
insulin-like growth factor binding protein 3 |
| chr2_+_161993465 | 0.28 |
ENST00000457476.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr5_+_81575281 | 0.28 |
ENST00000380167.4 |
ATP6AP1L |
ATPase, H+ transporting, lysosomal accessory protein 1-like |
| chr5_+_140762268 | 0.27 |
ENST00000518325.1 |
PCDHGA7 |
protocadherin gamma subfamily A, 7 |
| chr7_+_13141097 | 0.26 |
ENST00000411542.1 |
AC011288.2 |
AC011288.2 |
| chr9_-_99540328 | 0.25 |
ENST00000223428.4 ENST00000375231.1 ENST00000374641.3 |
ZNF510 |
zinc finger protein 510 |
| chr19_-_44388116 | 0.25 |
ENST00000587539.1 |
ZNF404 |
zinc finger protein 404 |
| chr5_-_88119580 | 0.25 |
ENST00000539796.1 |
MEF2C |
myocyte enhancer factor 2C |
| chr10_-_49860525 | 0.25 |
ENST00000435790.2 |
ARHGAP22 |
Rho GTPase activating protein 22 |
| chr10_-_27529486 | 0.24 |
ENST00000375888.1 |
ACBD5 |
acyl-CoA binding domain containing 5 |
| chr10_-_28623368 | 0.23 |
ENST00000441595.2 |
MPP7 |
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr18_-_59274139 | 0.23 |
ENST00000586949.1 |
RP11-879F14.2 |
RP11-879F14.2 |
| chr10_-_28571015 | 0.22 |
ENST00000375719.3 ENST00000375732.1 |
MPP7 |
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr7_+_134528635 | 0.22 |
ENST00000445569.2 |
CALD1 |
caldesmon 1 |
| chr19_+_52873166 | 0.22 |
ENST00000424032.2 ENST00000600321.1 ENST00000344085.5 ENST00000597976.1 ENST00000422689.2 |
ZNF880 |
zinc finger protein 880 |
| chr4_-_70725856 | 0.21 |
ENST00000226444.3 |
SULT1E1 |
sulfotransferase family 1E, estrogen-preferring, member 1 |
| chr2_+_161993412 | 0.21 |
ENST00000259075.2 ENST00000432002.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr9_+_18474204 | 0.21 |
ENST00000276935.6 |
ADAMTSL1 |
ADAMTS-like 1 |
| chrX_-_102531717 | 0.21 |
ENST00000372680.1 |
TCEAL5 |
transcription elongation factor A (SII)-like 5 |
| chr15_-_20193370 | 0.20 |
ENST00000558565.2 |
IGHV3OR15-7 |
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr16_-_66583701 | 0.20 |
ENST00000527800.1 ENST00000525974.1 ENST00000563369.2 |
TK2 |
thymidine kinase 2, mitochondrial |
| chr7_-_14026063 | 0.20 |
ENST00000443608.1 ENST00000438956.1 |
ETV1 |
ets variant 1 |
| chr6_-_152639479 | 0.19 |
ENST00000356820.4 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
| chr5_-_125930929 | 0.19 |
ENST00000553117.1 ENST00000447989.2 ENST00000409134.3 |
ALDH7A1 |
aldehyde dehydrogenase 7 family, member A1 |
| chr10_+_64133934 | 0.19 |
ENST00000395254.3 ENST00000395255.3 ENST00000410046.3 |
ZNF365 |
zinc finger protein 365 |
| chrX_+_154113317 | 0.18 |
ENST00000354461.2 |
H2AFB1 |
H2A histone family, member B1 |
| chr3_+_151591422 | 0.18 |
ENST00000362032.5 |
SUCNR1 |
succinate receptor 1 |
| chrX_-_154689596 | 0.18 |
ENST00000369444.2 |
H2AFB3 |
H2A histone family, member B3 |
| chr1_-_54411255 | 0.18 |
ENST00000371377.3 |
HSPB11 |
heat shock protein family B (small), member 11 |
| chr13_+_24144796 | 0.18 |
ENST00000403372.2 |
TNFRSF19 |
tumor necrosis factor receptor superfamily, member 19 |
| chr1_+_115572415 | 0.17 |
ENST00000256592.1 |
TSHB |
thyroid stimulating hormone, beta |
| chr6_+_114178512 | 0.17 |
ENST00000368635.4 |
MARCKS |
myristoylated alanine-rich protein kinase C substrate |
| chr5_+_115177178 | 0.17 |
ENST00000316788.7 |
AP3S1 |
adaptor-related protein complex 3, sigma 1 subunit |
| chr3_+_138340067 | 0.16 |
ENST00000479848.1 |
FAIM |
Fas apoptotic inhibitory molecule |
| chr4_-_138453606 | 0.16 |
ENST00000412923.2 ENST00000344876.4 ENST00000507846.1 ENST00000510305.1 |
PCDH18 |
protocadherin 18 |
| chr11_-_58612168 | 0.16 |
ENST00000287275.1 |
GLYATL2 |
glycine-N-acyltransferase-like 2 |
| chr12_+_8309630 | 0.16 |
ENST00000396570.3 |
ZNF705A |
zinc finger protein 705A |
| chr11_+_12399071 | 0.16 |
ENST00000539723.1 ENST00000550549.1 |
PARVA |
parvin, alpha |
| chrX_-_106243451 | 0.16 |
ENST00000355610.4 ENST00000535534.1 |
MORC4 |
MORC family CW-type zinc finger 4 |
| chr13_-_49107303 | 0.16 |
ENST00000344532.3 |
RCBTB2 |
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr3_-_141747950 | 0.16 |
ENST00000497579.1 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr6_-_52859968 | 0.15 |
ENST00000370959.1 |
GSTA4 |
glutathione S-transferase alpha 4 |
| chr3_+_16216137 | 0.15 |
ENST00000339732.5 |
GALNT15 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 15 |
| chr17_+_6918354 | 0.15 |
ENST00000552775.1 |
C17orf49 |
chromosome 17 open reading frame 49 |
| chr17_-_5321549 | 0.15 |
ENST00000572809.1 |
NUP88 |
nucleoporin 88kDa |
| chr16_+_16434185 | 0.15 |
ENST00000524823.2 |
AC138969.4 |
Protein PKD1P1 |
| chr12_+_26164645 | 0.15 |
ENST00000542004.1 |
RASSF8 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr2_-_231860596 | 0.15 |
ENST00000441063.1 ENST00000434094.1 ENST00000418330.1 ENST00000457803.1 ENST00000414876.1 ENST00000446741.1 ENST00000426904.1 |
AC105344.2 |
SPATA3 antisense RNA 1 (head to head) |
| chr7_-_140482926 | 0.14 |
ENST00000496384.2 |
BRAF |
v-raf murine sarcoma viral oncogene homolog B |
| chr12_-_42631529 | 0.14 |
ENST00000548917.1 |
YAF2 |
YY1 associated factor 2 |
| chrX_+_154610428 | 0.14 |
ENST00000354514.4 |
H2AFB2 |
H2A histone family, member B2 |
| chr3_+_138340049 | 0.14 |
ENST00000464668.1 |
FAIM |
Fas apoptotic inhibitory molecule |
| chr16_-_66584059 | 0.14 |
ENST00000417693.3 ENST00000544898.1 ENST00000569718.1 ENST00000527284.1 ENST00000299697.7 ENST00000451102.2 |
TK2 |
thymidine kinase 2, mitochondrial |
| chr2_-_152118352 | 0.14 |
ENST00000331426.5 |
RBM43 |
RNA binding motif protein 43 |
| chr20_+_1099233 | 0.13 |
ENST00000246015.4 ENST00000335877.6 ENST00000438768.2 |
PSMF1 |
proteasome (prosome, macropain) inhibitor subunit 1 (PI31) |
| chr17_+_6918093 | 0.13 |
ENST00000439424.2 |
C17orf49 |
chromosome 17 open reading frame 49 |
| chr19_-_35264089 | 0.13 |
ENST00000588760.1 ENST00000329285.8 ENST00000587354.2 |
ZNF599 |
zinc finger protein 599 |
| chrX_-_102942961 | 0.13 |
ENST00000434230.1 ENST00000418819.1 ENST00000360458.1 |
MORF4L2 |
mortality factor 4 like 2 |
| chr10_+_17270214 | 0.13 |
ENST00000544301.1 |
VIM |
vimentin |
| chr20_+_5986727 | 0.13 |
ENST00000378863.4 |
CRLS1 |
cardiolipin synthase 1 |
| chr1_+_8378140 | 0.13 |
ENST00000377479.2 |
SLC45A1 |
solute carrier family 45, member 1 |
| chr20_+_5986756 | 0.12 |
ENST00000452938.1 |
CRLS1 |
cardiolipin synthase 1 |
| chr1_+_212965170 | 0.12 |
ENST00000532324.1 ENST00000366974.4 ENST00000530441.1 ENST00000526641.1 ENST00000531963.1 ENST00000366973.4 ENST00000526997.1 ENST00000488246.2 |
TATDN3 |
TatD DNase domain containing 3 |
| chr10_+_5135981 | 0.12 |
ENST00000380554.3 |
AKR1C3 |
aldo-keto reductase family 1, member C3 |
| chr11_-_327537 | 0.12 |
ENST00000602735.1 |
IFITM3 |
interferon induced transmembrane protein 3 |
| chr4_+_175839506 | 0.12 |
ENST00000505141.1 ENST00000359240.3 ENST00000445694.1 |
ADAM29 |
ADAM metallopeptidase domain 29 |
| chr6_+_111408698 | 0.12 |
ENST00000368851.5 |
SLC16A10 |
solute carrier family 16 (aromatic amino acid transporter), member 10 |
| chr5_+_154320623 | 0.12 |
ENST00000523037.1 ENST00000265229.8 ENST00000439747.3 ENST00000522038.1 |
MRPL22 |
mitochondrial ribosomal protein L22 |
| chr4_+_175839551 | 0.11 |
ENST00000404450.4 ENST00000514159.1 |
ADAM29 |
ADAM metallopeptidase domain 29 |
| chr4_+_186990298 | 0.11 |
ENST00000296795.3 ENST00000513189.1 |
TLR3 |
toll-like receptor 3 |
| chrX_-_102943022 | 0.11 |
ENST00000433176.2 |
MORF4L2 |
mortality factor 4 like 2 |
| chr7_-_22862406 | 0.11 |
ENST00000372879.4 |
TOMM7 |
translocase of outer mitochondrial membrane 7 homolog (yeast) |
| chr1_-_151804314 | 0.11 |
ENST00000318247.6 |
RORC |
RAR-related orphan receptor C |
| chr2_-_45795145 | 0.11 |
ENST00000535761.1 |
SRBD1 |
S1 RNA binding domain 1 |
| chr17_-_40950698 | 0.10 |
ENST00000328434.7 |
COA3 |
cytochrome c oxidase assembly factor 3 |
| chr10_+_52152766 | 0.10 |
ENST00000596442.1 |
AC069547.2 |
Uncharacterized protein |
| chr4_+_3344141 | 0.10 |
ENST00000306648.7 |
RGS12 |
regulator of G-protein signaling 12 |
| chr11_+_110300607 | 0.10 |
ENST00000260270.2 |
FDX1 |
ferredoxin 1 |
| chr6_+_28092338 | 0.10 |
ENST00000340487.4 |
ZSCAN16 |
zinc finger and SCAN domain containing 16 |
| chr6_-_26235206 | 0.10 |
ENST00000244534.5 |
HIST1H1D |
histone cluster 1, H1d |
| chr6_+_140175987 | 0.10 |
ENST00000414038.1 ENST00000431609.1 |
RP5-899B16.1 |
RP5-899B16.1 |
| chr20_+_11008408 | 0.10 |
ENST00000378252.1 |
C20orf187 |
chromosome 20 open reading frame 187 |
| chr9_-_21482312 | 0.10 |
ENST00000448696.3 |
IFNE |
interferon, epsilon |
| chr6_-_134861089 | 0.10 |
ENST00000606039.1 |
RP11-557H15.4 |
RP11-557H15.4 |
| chr3_+_141457105 | 0.10 |
ENST00000480908.1 ENST00000393000.3 |
RNF7 |
ring finger protein 7 |
| chr7_-_22862448 | 0.10 |
ENST00000358435.4 |
TOMM7 |
translocase of outer mitochondrial membrane 7 homolog (yeast) |
| chr1_+_192127578 | 0.10 |
ENST00000367460.3 |
RGS18 |
regulator of G-protein signaling 18 |
| chr6_-_146057144 | 0.09 |
ENST00000367519.3 |
EPM2A |
epilepsy, progressive myoclonus type 2A, Lafora disease (laforin) |
| chr20_+_5987890 | 0.09 |
ENST00000378868.4 |
CRLS1 |
cardiolipin synthase 1 |
| chr9_+_125133315 | 0.09 |
ENST00000223423.4 ENST00000362012.2 |
PTGS1 |
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr12_-_7656357 | 0.09 |
ENST00000396620.3 ENST00000432237.2 ENST00000359156.4 |
CD163 |
CD163 molecule |
| chr7_-_144435985 | 0.09 |
ENST00000549981.1 |
TPK1 |
thiamin pyrophosphokinase 1 |
| chr1_-_212965104 | 0.09 |
ENST00000422588.2 ENST00000366975.6 ENST00000366977.3 ENST00000366976.1 |
NSL1 |
NSL1, MIS12 kinetochore complex component |
| chr13_-_70682590 | 0.09 |
ENST00000377844.4 |
KLHL1 |
kelch-like family member 1 |
| chr1_-_53608289 | 0.09 |
ENST00000371491.4 |
SLC1A7 |
solute carrier family 1 (glutamate transporter), member 7 |
| chr15_+_64386261 | 0.09 |
ENST00000560829.1 |
SNX1 |
sorting nexin 1 |
| chr12_+_12510352 | 0.09 |
ENST00000298571.6 |
LOH12CR1 |
loss of heterozygosity, 12, chromosomal region 1 |
| chr3_+_38537960 | 0.09 |
ENST00000453767.1 |
EXOG |
endo/exonuclease (5'-3'), endonuclease G-like |
| chr4_-_120243545 | 0.09 |
ENST00000274024.3 |
FABP2 |
fatty acid binding protein 2, intestinal |
| chr5_+_111755280 | 0.09 |
ENST00000600409.1 |
EPB41L4A-AS2 |
EPB41L4A antisense RNA 2 (head to head) |
| chr3_-_141719195 | 0.09 |
ENST00000397991.4 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr2_+_210517895 | 0.08 |
ENST00000447185.1 |
MAP2 |
microtubule-associated protein 2 |
| chr11_-_26593677 | 0.08 |
ENST00000527569.1 |
MUC15 |
mucin 15, cell surface associated |
| chr22_+_20105259 | 0.08 |
ENST00000416427.1 ENST00000421656.1 ENST00000423859.1 ENST00000418705.2 |
RANBP1 |
RAN binding protein 1 |
| chr12_+_113796347 | 0.08 |
ENST00000545182.2 ENST00000280800.3 |
PLBD2 |
phospholipase B domain containing 2 |
| chr1_+_112938803 | 0.08 |
ENST00000271277.6 |
CTTNBP2NL |
CTTNBP2 N-terminal like |
| chr6_+_47749718 | 0.08 |
ENST00000489301.2 ENST00000371211.2 ENST00000393699.2 |
OPN5 |
opsin 5 |
| chr8_-_145754428 | 0.08 |
ENST00000527462.1 ENST00000313465.5 ENST00000524821.1 |
C8orf82 |
chromosome 8 open reading frame 82 |
| chr4_-_138453559 | 0.08 |
ENST00000511115.1 |
PCDH18 |
protocadherin 18 |
| chr12_-_7596735 | 0.08 |
ENST00000416109.2 ENST00000396630.1 ENST00000313599.3 |
CD163L1 |
CD163 molecule-like 1 |
| chr6_+_154360476 | 0.08 |
ENST00000428397.2 |
OPRM1 |
opioid receptor, mu 1 |
| chr8_-_57123815 | 0.08 |
ENST00000316981.3 ENST00000423799.2 ENST00000429357.2 |
PLAG1 |
pleiomorphic adenoma gene 1 |
| chr7_+_138915102 | 0.08 |
ENST00000486663.1 |
UBN2 |
ubinuclein 2 |
| chr6_+_158733692 | 0.08 |
ENST00000367094.2 ENST00000367097.3 |
TULP4 |
tubby like protein 4 |
| chr9_-_116065551 | 0.08 |
ENST00000297894.5 |
RNF183 |
ring finger protein 183 |
| chr20_-_29978383 | 0.08 |
ENST00000339144.3 ENST00000376321.3 |
DEFB119 |
defensin, beta 119 |
| chr7_-_141957847 | 0.07 |
ENST00000552471.1 ENST00000547058.2 |
PRSS58 |
protease, serine, 58 |
| chr10_-_50396407 | 0.07 |
ENST00000374153.2 ENST00000374151.3 |
C10orf128 |
chromosome 10 open reading frame 128 |
| chr6_+_26104104 | 0.07 |
ENST00000377803.2 |
HIST1H4C |
histone cluster 1, H4c |
| chr14_-_106926724 | 0.07 |
ENST00000434710.1 |
IGHV3-43 |
immunoglobulin heavy variable 3-43 |
| chr7_-_14026123 | 0.07 |
ENST00000420159.2 ENST00000399357.3 ENST00000403527.1 |
ETV1 |
ets variant 1 |
| chrX_+_43515467 | 0.07 |
ENST00000338702.3 ENST00000542639.1 |
MAOA |
monoamine oxidase A |
| chr17_+_29664830 | 0.07 |
ENST00000444181.2 ENST00000417592.2 |
NF1 |
neurofibromin 1 |
| chr1_-_228613026 | 0.07 |
ENST00000366696.1 |
HIST3H3 |
histone cluster 3, H3 |
| chr11_+_55578854 | 0.07 |
ENST00000333973.2 |
OR5L1 |
olfactory receptor, family 5, subfamily L, member 1 |
| chr9_-_85882145 | 0.07 |
ENST00000328788.1 |
FRMD3 |
FERM domain containing 3 |
| chr19_+_36139125 | 0.07 |
ENST00000246554.3 |
COX6B1 |
cytochrome c oxidase subunit VIb polypeptide 1 (ubiquitous) |
| chr8_-_79470728 | 0.07 |
ENST00000522807.1 ENST00000519242.1 ENST00000522302.1 |
RP11-594N15.2 |
RP11-594N15.2 |
| chr10_-_75226166 | 0.07 |
ENST00000544628.1 |
PPP3CB |
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr4_+_113568207 | 0.06 |
ENST00000511529.1 |
LARP7 |
La ribonucleoprotein domain family, member 7 |
| chr11_-_117186946 | 0.06 |
ENST00000313005.6 ENST00000528053.1 |
BACE1 |
beta-site APP-cleaving enzyme 1 |
| chr8_-_101117847 | 0.06 |
ENST00000523287.1 ENST00000519092.1 |
RGS22 |
regulator of G-protein signaling 22 |
| chr19_+_13842559 | 0.06 |
ENST00000586600.1 |
CCDC130 |
coiled-coil domain containing 130 |
| chr19_-_4535233 | 0.06 |
ENST00000381848.3 ENST00000588887.1 ENST00000586133.1 |
PLIN5 |
perilipin 5 |
| chr4_+_74347400 | 0.06 |
ENST00000226355.3 |
AFM |
afamin |
| chr17_+_79849872 | 0.06 |
ENST00000584197.1 ENST00000583839.1 |
ANAPC11 |
anaphase promoting complex subunit 11 |
| chr18_+_616672 | 0.06 |
ENST00000338387.7 |
CLUL1 |
clusterin-like 1 (retinal) |
| chr4_+_169552748 | 0.06 |
ENST00000504519.1 ENST00000512127.1 |
PALLD |
palladin, cytoskeletal associated protein |
| chr14_+_55493920 | 0.06 |
ENST00000395472.2 ENST00000555846.1 |
SOCS4 |
suppressor of cytokine signaling 4 |
| chr9_-_28670283 | 0.06 |
ENST00000379992.2 |
LINGO2 |
leucine rich repeat and Ig domain containing 2 |
| chr4_+_190861993 | 0.06 |
ENST00000524583.1 ENST00000531991.2 |
FRG1 |
FSHD region gene 1 |
| chr10_-_14050522 | 0.06 |
ENST00000342409.2 |
FRMD4A |
FERM domain containing 4A |
| chr21_-_43735628 | 0.06 |
ENST00000291525.10 ENST00000518498.1 |
TFF3 |
trefoil factor 3 (intestinal) |
| chr1_+_168250194 | 0.06 |
ENST00000367821.3 |
TBX19 |
T-box 19 |
| chr1_+_233086326 | 0.06 |
ENST00000366628.5 ENST00000366627.4 |
NTPCR |
nucleoside-triphosphatase, cancer-related |
| chr5_+_140588269 | 0.05 |
ENST00000541609.1 ENST00000239450.2 |
PCDHB12 |
protocadherin beta 12 |
| chr11_+_71544246 | 0.05 |
ENST00000328698.1 |
DEFB108B |
defensin, beta 108B |
| chr1_+_76540386 | 0.05 |
ENST00000328299.3 |
ST6GALNAC3 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
| chr5_-_36301984 | 0.05 |
ENST00000502994.1 ENST00000515759.1 ENST00000296604.3 |
RANBP3L |
RAN binding protein 3-like |
| chr3_+_136649311 | 0.05 |
ENST00000469404.1 ENST00000467911.1 |
NCK1 |
NCK adaptor protein 1 |
| chr18_+_45778672 | 0.05 |
ENST00000600091.1 |
AC091150.1 |
HCG1818186; Uncharacterized protein |
| chr10_+_116697946 | 0.05 |
ENST00000298746.3 |
TRUB1 |
TruB pseudouridine (psi) synthase family member 1 |
| chr20_+_31870927 | 0.05 |
ENST00000253354.1 |
BPIFB1 |
BPI fold containing family B, member 1 |
| chr7_-_76829125 | 0.05 |
ENST00000248598.5 |
FGL2 |
fibrinogen-like 2 |
| chr11_-_26593649 | 0.05 |
ENST00000455601.2 |
MUC15 |
mucin 15, cell surface associated |
| chr15_-_75748115 | 0.05 |
ENST00000360439.4 |
SIN3A |
SIN3 transcription regulator family member A |
| chr12_+_64798095 | 0.05 |
ENST00000332707.5 |
XPOT |
exportin, tRNA |
| chr11_-_89224488 | 0.05 |
ENST00000534731.1 ENST00000527626.1 |
NOX4 |
NADPH oxidase 4 |
| chr20_+_3776371 | 0.05 |
ENST00000245960.5 |
CDC25B |
cell division cycle 25B |
| chr2_-_27712583 | 0.05 |
ENST00000260570.3 ENST00000359466.6 ENST00000416524.2 |
IFT172 |
intraflagellar transport 172 homolog (Chlamydomonas) |
| chr21_-_22175341 | 0.05 |
ENST00000416768.1 ENST00000452561.1 ENST00000419299.1 ENST00000437238.1 |
LINC00320 |
long intergenic non-protein coding RNA 320 |
| chr15_-_56757329 | 0.04 |
ENST00000260453.3 |
MNS1 |
meiosis-specific nuclear structural 1 |
| chr12_-_22063787 | 0.04 |
ENST00000544039.1 |
ABCC9 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr4_-_159956333 | 0.04 |
ENST00000434826.2 |
C4orf45 |
chromosome 4 open reading frame 45 |
| chr6_-_89927151 | 0.04 |
ENST00000454853.2 |
GABRR1 |
gamma-aminobutyric acid (GABA) A receptor, rho 1 |
| chr14_-_80697396 | 0.04 |
ENST00000557010.1 |
DIO2 |
deiodinase, iodothyronine, type II |
| chr17_+_1944790 | 0.04 |
ENST00000575162.1 |
DPH1 |
diphthamide biosynthesis 1 |
| chr5_+_147691979 | 0.04 |
ENST00000274565.4 |
SPINK7 |
serine peptidase inhibitor, Kazal type 7 (putative) |
| chr11_-_62911693 | 0.04 |
ENST00000417740.1 ENST00000326192.5 |
SLC22A24 |
solute carrier family 22, member 24 |
| chr6_+_26440700 | 0.04 |
ENST00000494393.1 ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3 |
butyrophilin, subfamily 3, member A3 |
| chr5_+_140593509 | 0.04 |
ENST00000341948.4 |
PCDHB13 |
protocadherin beta 13 |
| chrX_-_77225135 | 0.04 |
ENST00000458128.1 |
PGAM4 |
phosphoglycerate mutase family member 4 |
| chr2_+_67624430 | 0.04 |
ENST00000272342.5 |
ETAA1 |
Ewing tumor-associated antigen 1 |
| chr4_-_103746683 | 0.04 |
ENST00000504211.1 ENST00000508476.1 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chr12_+_131438443 | 0.04 |
ENST00000261654.5 |
GPR133 |
G protein-coupled receptor 133 |
| chr4_+_86525299 | 0.04 |
ENST00000512201.1 |
ARHGAP24 |
Rho GTPase activating protein 24 |
| chr1_+_152943122 | 0.04 |
ENST00000328051.2 |
SPRR4 |
small proline-rich protein 4 |
| chr7_+_138943265 | 0.04 |
ENST00000483726.1 |
UBN2 |
ubinuclein 2 |
| chr11_-_18548426 | 0.04 |
ENST00000357193.3 ENST00000536719.1 |
TSG101 |
tumor susceptibility 101 |
| chr6_-_25874440 | 0.04 |
ENST00000361703.6 ENST00000397060.4 |
SLC17A3 |
solute carrier family 17 (organic anion transporter), member 3 |
| chrX_+_84258832 | 0.04 |
ENST00000373173.2 |
APOOL |
apolipoprotein O-like |
| chr17_+_74463650 | 0.04 |
ENST00000392492.3 |
AANAT |
aralkylamine N-acetyltransferase |
| chr9_-_215744 | 0.04 |
ENST00000382387.2 |
C9orf66 |
chromosome 9 open reading frame 66 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.5 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 0.9 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.1 | 0.3 | GO:0046125 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.1 | 0.5 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.4 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.1 | 0.2 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.1 | 0.2 | GO:0031456 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.1 | 0.2 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 0.6 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 0.4 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.1 | 0.5 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.7 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.0 | GO:0051031 | tRNA export from nucleus(GO:0006409) tRNA transport(GO:0051031) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.3 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0034343 | microglial cell activation involved in immune response(GO:0002282) type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.1 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.0 | 0.1 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.0 | 0.6 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.3 | GO:0018394 | histone acetylation(GO:0016573) internal peptidyl-lysine acetylation(GO:0018393) peptidyl-lysine acetylation(GO:0018394) |
| 0.0 | 0.2 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.2 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.4 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.2 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.1 | GO:0061358 | negative regulation of Wnt protein secretion(GO:0061358) |
| 0.0 | 0.1 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.2 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.0 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.0 | 0.0 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.0 | 0.0 | GO:2000397 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.3 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.0 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.2 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.0 | 0.0 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.5 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.1 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 0.0 | 0.0 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 0.0 | 0.1 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.1 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.0 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 | 0.3 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.0 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.1 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.2 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.3 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.5 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.0 | 0.4 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.3 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.0 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.2 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.1 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.2 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 0.3 | GO:0008808 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.1 | 0.3 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.1 | 0.6 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.1 | 0.5 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 0.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.3 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.2 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0047718 | androsterone dehydrogenase activity(GO:0047023) indanol dehydrogenase activity(GO:0047718) |
| 0.0 | 0.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.1 | GO:0038047 | beta-endorphin receptor activity(GO:0004979) morphine receptor activity(GO:0038047) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.2 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.0 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.1 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.1 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 0.0 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.0 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.6 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.3 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |