Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for POU6F2
Z-value: 0.77
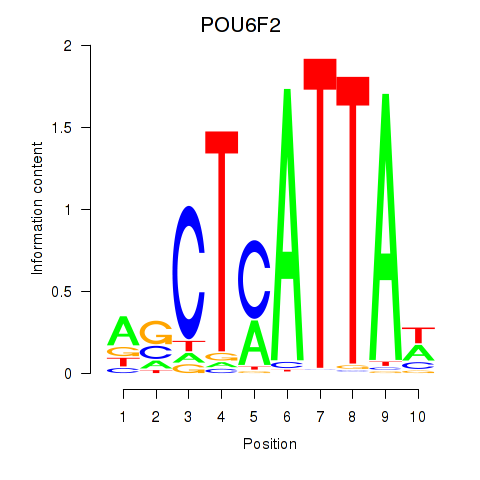
Transcription factors associated with POU6F2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU6F2
|
ENSG00000106536.15 | POU6F2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU6F2 | hg19_v2_chr7_+_39125365_39125489, hg19_v2_chr7_+_39017504_39017598 | -0.34 | 4.2e-01 | Click! |
Activity profile of POU6F2 motif
Sorted Z-values of POU6F2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of POU6F2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_158300556 | 0.62 |
ENST00000264192.3 |
CYTIP |
cytohesin 1 interacting protein |
| chr8_-_42234745 | 0.33 |
ENST00000220812.2 |
DKK4 |
dickkopf WNT signaling pathway inhibitor 4 |
| chr17_+_37784749 | 0.32 |
ENST00000394265.1 ENST00000394267.2 |
PPP1R1B |
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr1_-_149900122 | 0.26 |
ENST00000271628.8 |
SF3B4 |
splicing factor 3b, subunit 4, 49kDa |
| chr4_-_66536057 | 0.22 |
ENST00000273854.3 |
EPHA5 |
EPH receptor A5 |
| chr3_-_33686925 | 0.21 |
ENST00000485378.2 ENST00000313350.6 ENST00000487200.1 |
CLASP2 |
cytoplasmic linker associated protein 2 |
| chr5_-_156486120 | 0.20 |
ENST00000522693.1 |
HAVCR1 |
hepatitis A virus cellular receptor 1 |
| chr12_-_52946923 | 0.19 |
ENST00000267119.5 |
KRT71 |
keratin 71 |
| chr13_-_110438914 | 0.18 |
ENST00000375856.3 |
IRS2 |
insulin receptor substrate 2 |
| chrX_-_38186811 | 0.18 |
ENST00000318842.7 |
RPGR |
retinitis pigmentosa GTPase regulator |
| chr16_+_53133070 | 0.17 |
ENST00000565832.1 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
| chr14_-_36988882 | 0.17 |
ENST00000498187.2 |
NKX2-1 |
NK2 homeobox 1 |
| chrX_-_38186775 | 0.17 |
ENST00000339363.3 ENST00000309513.3 ENST00000338898.3 ENST00000342811.3 ENST00000378505.2 |
RPGR |
retinitis pigmentosa GTPase regulator |
| chr6_+_130339710 | 0.17 |
ENST00000526087.1 ENST00000533560.1 ENST00000361794.2 |
L3MBTL3 |
l(3)mbt-like 3 (Drosophila) |
| chr1_-_193028632 | 0.16 |
ENST00000421683.1 |
UCHL5 |
ubiquitin carboxyl-terminal hydrolase L5 |
| chr7_+_141478242 | 0.16 |
ENST00000247881.2 |
TAS2R4 |
taste receptor, type 2, member 4 |
| chr3_-_33686743 | 0.16 |
ENST00000333778.6 ENST00000539981.1 |
CLASP2 |
cytoplasmic linker associated protein 2 |
| chr2_-_182545603 | 0.15 |
ENST00000295108.3 |
NEUROD1 |
neuronal differentiation 1 |
| chr1_-_193028426 | 0.15 |
ENST00000367450.3 ENST00000530098.2 ENST00000367451.4 ENST00000367448.1 ENST00000367449.1 |
UCHL5 |
ubiquitin carboxyl-terminal hydrolase L5 |
| chr2_+_169757750 | 0.14 |
ENST00000375363.3 ENST00000429379.2 ENST00000421979.1 |
G6PC2 |
glucose-6-phosphatase, catalytic, 2 |
| chr14_-_69261310 | 0.14 |
ENST00000336440.3 |
ZFP36L1 |
ZFP36 ring finger protein-like 1 |
| chr20_+_56136136 | 0.13 |
ENST00000319441.4 ENST00000543666.1 |
PCK1 |
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr1_+_107683436 | 0.13 |
ENST00000370068.1 |
NTNG1 |
netrin G1 |
| chr12_+_54348618 | 0.13 |
ENST00000243103.3 |
HOXC12 |
homeobox C12 |
| chr1_-_193028621 | 0.13 |
ENST00000367455.4 ENST00000367454.1 |
UCHL5 |
ubiquitin carboxyl-terminal hydrolase L5 |
| chr10_+_94352956 | 0.13 |
ENST00000260731.3 |
KIF11 |
kinesin family member 11 |
| chr17_+_18086392 | 0.12 |
ENST00000541285.1 |
ALKBH5 |
alkB, alkylation repair homolog 5 (E. coli) |
| chr11_-_27494309 | 0.12 |
ENST00000389858.4 |
LGR4 |
leucine-rich repeat containing G protein-coupled receptor 4 |
| chrX_-_18690210 | 0.12 |
ENST00000379984.3 |
RS1 |
retinoschisin 1 |
| chr15_+_75080883 | 0.12 |
ENST00000567571.1 |
CSK |
c-src tyrosine kinase |
| chr11_-_27494279 | 0.11 |
ENST00000379214.4 |
LGR4 |
leucine-rich repeat containing G protein-coupled receptor 4 |
| chr1_+_107683644 | 0.11 |
ENST00000370067.1 |
NTNG1 |
netrin G1 |
| chr3_+_141106458 | 0.11 |
ENST00000509883.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr9_-_3469181 | 0.11 |
ENST00000366116.2 |
AL365202.1 |
Uncharacterized protein |
| chr1_+_10271674 | 0.11 |
ENST00000377086.1 |
KIF1B |
kinesin family member 1B |
| chr3_+_189507460 | 0.11 |
ENST00000434928.1 |
TP63 |
tumor protein p63 |
| chr6_+_25754927 | 0.11 |
ENST00000377905.4 ENST00000439485.2 |
SLC17A4 |
solute carrier family 17, member 4 |
| chr12_-_87232644 | 0.10 |
ENST00000549405.2 |
MGAT4C |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr4_-_164395014 | 0.10 |
ENST00000280605.3 |
TKTL2 |
transketolase-like 2 |
| chr4_-_74486217 | 0.10 |
ENST00000335049.5 ENST00000307439.5 |
RASSF6 |
Ras association (RalGDS/AF-6) domain family member 6 |
| chr4_-_22444733 | 0.10 |
ENST00000508133.1 |
GPR125 |
G protein-coupled receptor 125 |
| chr14_+_22984601 | 0.10 |
ENST00000390509.1 |
TRAJ28 |
T cell receptor alpha joining 28 |
| chr6_-_155635583 | 0.10 |
ENST00000367166.4 |
TFB1M |
transcription factor B1, mitochondrial |
| chr3_-_127541194 | 0.10 |
ENST00000453507.2 |
MGLL |
monoglyceride lipase |
| chr6_-_89927151 | 0.09 |
ENST00000454853.2 |
GABRR1 |
gamma-aminobutyric acid (GABA) A receptor, rho 1 |
| chr12_+_121131970 | 0.09 |
ENST00000535656.1 |
MLEC |
malectin |
| chr14_+_32798547 | 0.09 |
ENST00000557354.1 ENST00000557102.1 ENST00000557272.1 |
AKAP6 |
A kinase (PRKA) anchor protein 6 |
| chr15_+_96869165 | 0.09 |
ENST00000421109.2 |
NR2F2 |
nuclear receptor subfamily 2, group F, member 2 |
| chr1_-_208417620 | 0.09 |
ENST00000367033.3 |
PLXNA2 |
plexin A2 |
| chr4_+_110749143 | 0.09 |
ENST00000317735.4 |
RRH |
retinal pigment epithelium-derived rhodopsin homolog |
| chr5_+_159343688 | 0.09 |
ENST00000306675.3 |
ADRA1B |
adrenoceptor alpha 1B |
| chr15_+_93443419 | 0.09 |
ENST00000557381.1 ENST00000420239.2 |
CHD2 |
chromodomain helicase DNA binding protein 2 |
| chr12_-_30887948 | 0.08 |
ENST00000433722.2 |
CAPRIN2 |
caprin family member 2 |
| chr10_+_18549645 | 0.08 |
ENST00000396576.2 |
CACNB2 |
calcium channel, voltage-dependent, beta 2 subunit |
| chr8_+_26150628 | 0.08 |
ENST00000523925.1 ENST00000315985.7 |
PPP2R2A |
protein phosphatase 2, regulatory subunit B, alpha |
| chr5_-_139726181 | 0.08 |
ENST00000507104.1 ENST00000230990.6 |
HBEGF |
heparin-binding EGF-like growth factor |
| chr3_+_141105705 | 0.08 |
ENST00000513258.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr17_-_38911580 | 0.08 |
ENST00000312150.4 |
KRT25 |
keratin 25 |
| chr9_-_110251836 | 0.08 |
ENST00000374672.4 |
KLF4 |
Kruppel-like factor 4 (gut) |
| chr6_+_151646800 | 0.07 |
ENST00000354675.6 |
AKAP12 |
A kinase (PRKA) anchor protein 12 |
| chr1_+_62439037 | 0.07 |
ENST00000545929.1 |
INADL |
InaD-like (Drosophila) |
| chr6_-_87804815 | 0.07 |
ENST00000369582.2 |
CGA |
glycoprotein hormones, alpha polypeptide |
| chr7_+_115862858 | 0.07 |
ENST00000393481.2 |
TES |
testis derived transcript (3 LIM domains) |
| chr4_-_74486109 | 0.07 |
ENST00000395777.2 |
RASSF6 |
Ras association (RalGDS/AF-6) domain family member 6 |
| chr1_-_182641367 | 0.07 |
ENST00000508450.1 |
RGS8 |
regulator of G-protein signaling 8 |
| chr7_-_14028488 | 0.07 |
ENST00000405358.4 |
ETV1 |
ets variant 1 |
| chr7_-_73038867 | 0.07 |
ENST00000313375.3 ENST00000354613.1 ENST00000395189.1 ENST00000453275.1 |
MLXIPL |
MLX interacting protein-like |
| chr6_+_29274403 | 0.07 |
ENST00000377160.2 |
OR14J1 |
olfactory receptor, family 14, subfamily J, member 1 |
| chr1_-_234667504 | 0.07 |
ENST00000421207.1 ENST00000435574.1 |
RP5-855F14.1 |
RP5-855F14.1 |
| chr11_+_128563652 | 0.07 |
ENST00000527786.2 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr11_-_5364809 | 0.07 |
ENST00000300773.2 |
OR51B5 |
olfactory receptor, family 51, subfamily B, member 5 |
| chr10_-_94257512 | 0.07 |
ENST00000371581.5 |
IDE |
insulin-degrading enzyme |
| chr4_+_86525299 | 0.07 |
ENST00000512201.1 |
ARHGAP24 |
Rho GTPase activating protein 24 |
| chr14_+_32798462 | 0.07 |
ENST00000280979.4 |
AKAP6 |
A kinase (PRKA) anchor protein 6 |
| chrX_-_122756660 | 0.07 |
ENST00000441692.1 |
THOC2 |
THO complex 2 |
| chr7_-_73038822 | 0.06 |
ENST00000414749.2 ENST00000429400.2 ENST00000434326.1 |
MLXIPL |
MLX interacting protein-like |
| chr7_-_14029283 | 0.06 |
ENST00000433547.1 ENST00000405192.2 |
ETV1 |
ets variant 1 |
| chr13_+_98612446 | 0.06 |
ENST00000496368.1 ENST00000421861.2 ENST00000357602.3 |
IPO5 |
importin 5 |
| chr9_-_21482312 | 0.06 |
ENST00000448696.3 |
IFNE |
interferon, epsilon |
| chr4_-_74486347 | 0.06 |
ENST00000342081.3 |
RASSF6 |
Ras association (RalGDS/AF-6) domain family member 6 |
| chr15_-_54025300 | 0.06 |
ENST00000559418.1 |
WDR72 |
WD repeat domain 72 |
| chr1_-_2458026 | 0.06 |
ENST00000435556.3 ENST00000378466.3 |
PANK4 |
pantothenate kinase 4 |
| chr5_-_140998481 | 0.06 |
ENST00000518047.1 |
DIAPH1 |
diaphanous-related formin 1 |
| chr9_+_109625378 | 0.06 |
ENST00000277225.5 ENST00000457913.1 ENST00000472574.1 |
ZNF462 |
zinc finger protein 462 |
| chr17_-_27418537 | 0.06 |
ENST00000408971.2 |
TIAF1 |
TGFB1-induced anti-apoptotic factor 1 |
| chrX_-_110655306 | 0.06 |
ENST00000371993.2 |
DCX |
doublecortin |
| chr6_+_50786414 | 0.06 |
ENST00000344788.3 ENST00000393655.3 ENST00000263046.4 |
TFAP2B |
transcription factor AP-2 beta (activating enhancer binding protein 2 beta) |
| chrX_+_10031499 | 0.06 |
ENST00000454666.1 |
WWC3 |
WWC family member 3 |
| chr2_-_105030466 | 0.06 |
ENST00000449772.1 |
AC068535.3 |
AC068535.3 |
| chr3_-_45838011 | 0.05 |
ENST00000358525.4 ENST00000413781.1 |
SLC6A20 |
solute carrier family 6 (proline IMINO transporter), member 20 |
| chr20_+_11008408 | 0.05 |
ENST00000378252.1 |
C20orf187 |
chromosome 20 open reading frame 187 |
| chr2_-_61697862 | 0.05 |
ENST00000398571.2 |
USP34 |
ubiquitin specific peptidase 34 |
| chr8_+_92261516 | 0.05 |
ENST00000276609.3 ENST00000309536.2 |
SLC26A7 |
solute carrier family 26 (anion exchanger), member 7 |
| chr8_+_77593474 | 0.05 |
ENST00000455469.2 ENST00000050961.6 |
ZFHX4 |
zinc finger homeobox 4 |
| chr8_+_77593448 | 0.05 |
ENST00000521891.2 |
ZFHX4 |
zinc finger homeobox 4 |
| chr3_+_189507523 | 0.05 |
ENST00000437221.1 ENST00000392463.2 ENST00000392461.3 ENST00000449992.1 ENST00000456148.1 |
TP63 |
tumor protein p63 |
| chr21_-_32253874 | 0.04 |
ENST00000332378.4 |
KRTAP11-1 |
keratin associated protein 11-1 |
| chr4_+_78829479 | 0.04 |
ENST00000504901.1 |
MRPL1 |
mitochondrial ribosomal protein L1 |
| chr8_-_90996837 | 0.04 |
ENST00000519426.1 ENST00000265433.3 |
NBN |
nibrin |
| chr11_+_125034586 | 0.04 |
ENST00000298282.9 |
PKNOX2 |
PBX/knotted 1 homeobox 2 |
| chr5_-_140998616 | 0.04 |
ENST00000389054.3 ENST00000398562.2 ENST00000389057.5 ENST00000398566.3 ENST00000398557.4 ENST00000253811.6 |
DIAPH1 |
diaphanous-related formin 1 |
| chr22_-_50524298 | 0.04 |
ENST00000311597.5 |
MLC1 |
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr22_-_50523760 | 0.04 |
ENST00000395876.2 |
MLC1 |
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr1_-_204329013 | 0.04 |
ENST00000272203.3 ENST00000414478.1 |
PLEKHA6 |
pleckstrin homology domain containing, family A member 6 |
| chr9_-_34729457 | 0.04 |
ENST00000378788.3 |
FAM205A |
family with sequence similarity 205, member A |
| chr11_-_63439013 | 0.04 |
ENST00000398868.3 |
ATL3 |
atlastin GTPase 3 |
| chr21_+_33671160 | 0.04 |
ENST00000303645.5 |
MRAP |
melanocortin 2 receptor accessory protein |
| chrX_-_30877837 | 0.04 |
ENST00000378930.3 |
TAB3 |
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr17_-_46716647 | 0.04 |
ENST00000608940.1 |
RP11-357H14.17 |
RP11-357H14.17 |
| chr10_+_77056134 | 0.04 |
ENST00000528121.1 ENST00000416398.1 |
ZNF503-AS1 |
ZNF503 antisense RNA 1 |
| chr17_-_39553844 | 0.04 |
ENST00000251645.2 |
KRT31 |
keratin 31 |
| chr2_-_69098566 | 0.04 |
ENST00000295379.1 |
BMP10 |
bone morphogenetic protein 10 |
| chr3_-_57199397 | 0.04 |
ENST00000296318.7 |
IL17RD |
interleukin 17 receptor D |
| chr5_-_111093167 | 0.04 |
ENST00000446294.2 ENST00000419114.2 |
NREP |
neuronal regeneration related protein |
| chr5_-_111093081 | 0.03 |
ENST00000453526.2 ENST00000509427.1 |
NREP |
neuronal regeneration related protein |
| chr7_-_80141328 | 0.03 |
ENST00000398291.3 |
GNAT3 |
guanine nucleotide binding protein, alpha transducing 3 |
| chr19_-_7698599 | 0.03 |
ENST00000311069.5 |
PCP2 |
Purkinje cell protein 2 |
| chr1_-_21606013 | 0.03 |
ENST00000357071.4 |
ECE1 |
endothelin converting enzyme 1 |
| chr21_-_31864275 | 0.03 |
ENST00000334063.4 |
KRTAP19-3 |
keratin associated protein 19-3 |
| chr14_+_39583427 | 0.03 |
ENST00000308317.6 ENST00000396249.2 ENST00000250379.8 ENST00000534684.2 ENST00000527381.1 |
GEMIN2 |
gem (nuclear organelle) associated protein 2 |
| chr9_-_13165457 | 0.03 |
ENST00000542239.1 ENST00000538841.1 ENST00000433359.2 |
MPDZ |
multiple PDZ domain protein |
| chr8_+_22424551 | 0.03 |
ENST00000523348.1 |
SORBS3 |
sorbin and SH3 domain containing 3 |
| chr3_+_189507432 | 0.03 |
ENST00000354600.5 |
TP63 |
tumor protein p63 |
| chr8_+_119294456 | 0.03 |
ENST00000366457.2 |
AC023590.1 |
Uncharacterized protein |
| chr3_-_51909600 | 0.03 |
ENST00000446461.1 |
IQCF5 |
IQ motif containing F5 |
| chr13_-_44735393 | 0.03 |
ENST00000400419.1 |
SMIM2 |
small integral membrane protein 2 |
| chr17_-_33390667 | 0.03 |
ENST00000378516.2 ENST00000268850.7 ENST00000394597.2 |
RFFL |
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr10_-_128210005 | 0.03 |
ENST00000284694.7 ENST00000454341.1 ENST00000432642.1 ENST00000392694.1 |
C10orf90 |
chromosome 10 open reading frame 90 |
| chr6_-_33160231 | 0.03 |
ENST00000395194.1 ENST00000457788.1 ENST00000341947.2 ENST00000357486.1 ENST00000374714.1 ENST00000374713.1 ENST00000395197.1 ENST00000374712.1 ENST00000361917.1 ENST00000374708.4 |
COL11A2 |
collagen, type XI, alpha 2 |
| chr1_+_193028552 | 0.03 |
ENST00000400968.2 ENST00000432079.1 |
TROVE2 |
TROVE domain family, member 2 |
| chr18_-_13915530 | 0.02 |
ENST00000327606.3 |
MC2R |
melanocortin 2 receptor (adrenocorticotropic hormone) |
| chr8_-_90769422 | 0.02 |
ENST00000524190.1 ENST00000523859.1 |
RP11-37B2.1 |
RP11-37B2.1 |
| chr14_-_27066960 | 0.02 |
ENST00000539517.2 |
NOVA1 |
neuro-oncological ventral antigen 1 |
| chr13_-_95131923 | 0.02 |
ENST00000377028.5 ENST00000446125.1 |
DCT |
dopachrome tautomerase |
| chr10_+_114710425 | 0.02 |
ENST00000352065.5 ENST00000369395.1 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr11_+_101918153 | 0.02 |
ENST00000434758.2 ENST00000526781.1 ENST00000534360.1 |
C11orf70 |
chromosome 11 open reading frame 70 |
| chr20_+_58515417 | 0.02 |
ENST00000360816.3 |
FAM217B |
family with sequence similarity 217, member B |
| chr4_+_175204818 | 0.02 |
ENST00000503780.1 |
CEP44 |
centrosomal protein 44kDa |
| chr15_+_58702742 | 0.02 |
ENST00000356113.6 ENST00000414170.3 |
LIPC |
lipase, hepatic |
| chr3_+_111717600 | 0.02 |
ENST00000273368.4 |
TAGLN3 |
transgelin 3 |
| chr17_-_39471947 | 0.02 |
ENST00000334202.3 |
KRTAP17-1 |
keratin associated protein 17-1 |
| chr7_-_83824169 | 0.02 |
ENST00000265362.4 |
SEMA3A |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr14_-_36989336 | 0.02 |
ENST00000522719.2 |
NKX2-1 |
NK2 homeobox 1 |
| chr4_+_66536248 | 0.02 |
ENST00000514260.1 ENST00000507117.1 |
RP11-807H7.1 |
RP11-807H7.1 |
| chr17_+_48243352 | 0.02 |
ENST00000344627.6 ENST00000262018.3 ENST00000543315.1 ENST00000451235.2 ENST00000511303.1 |
SGCA |
sarcoglycan, alpha (50kDa dystrophin-associated glycoprotein) |
| chr12_-_16761007 | 0.02 |
ENST00000354662.1 ENST00000441439.2 |
LMO3 |
LIM domain only 3 (rhombotin-like 2) |
| chr3_+_111717511 | 0.01 |
ENST00000478951.1 ENST00000393917.2 |
TAGLN3 |
transgelin 3 |
| chr4_+_40198527 | 0.01 |
ENST00000381799.5 |
RHOH |
ras homolog family member H |
| chr12_-_86230315 | 0.01 |
ENST00000361228.3 |
RASSF9 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
| chr7_-_5465045 | 0.01 |
ENST00000399434.2 |
TNRC18 |
trinucleotide repeat containing 18 |
| chrX_-_23926004 | 0.01 |
ENST00000379226.4 ENST00000379220.3 |
APOO |
apolipoprotein O |
| chr10_+_35484793 | 0.01 |
ENST00000488741.1 ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM |
cAMP responsive element modulator |
| chr9_+_131062367 | 0.01 |
ENST00000601297.1 |
AL359091.2 |
CDNA: FLJ21673 fis, clone COL09042; HCG2036511; Uncharacterized protein |
| chr5_-_134914673 | 0.01 |
ENST00000512158.1 |
CXCL14 |
chemokine (C-X-C motif) ligand 14 |
| chr19_-_7697857 | 0.01 |
ENST00000598935.1 |
PCP2 |
Purkinje cell protein 2 |
| chr17_-_10421853 | 0.01 |
ENST00000226207.5 |
MYH1 |
myosin, heavy chain 1, skeletal muscle, adult |
| chr17_+_12569306 | 0.01 |
ENST00000425538.1 |
MYOCD |
myocardin |
| chr7_+_5465382 | 0.01 |
ENST00000609130.1 |
RP11-1275H24.2 |
RP11-1275H24.2 |
| chr6_+_42123141 | 0.01 |
ENST00000418175.1 ENST00000541991.1 ENST00000053469.4 ENST00000394237.1 ENST00000372963.1 |
GUCA1A RP1-139D8.6 |
guanylate cyclase activator 1A (retina) RP1-139D8.6 |
| chr9_-_79520989 | 0.01 |
ENST00000376713.3 ENST00000376718.3 ENST00000428286.1 |
PRUNE2 |
prune homolog 2 (Drosophila) |
| chr10_-_48416849 | 0.01 |
ENST00000249598.1 |
GDF2 |
growth differentiation factor 2 |
| chr3_+_111718036 | 0.01 |
ENST00000455401.2 |
TAGLN3 |
transgelin 3 |
| chr3_-_101039402 | 0.01 |
ENST00000193391.7 |
IMPG2 |
interphotoreceptor matrix proteoglycan 2 |
| chr21_-_35899113 | 0.01 |
ENST00000492600.1 ENST00000481448.1 ENST00000381132.2 |
RCAN1 |
regulator of calcineurin 1 |
| chr1_+_175036966 | 0.01 |
ENST00000239462.4 |
TNN |
tenascin N |
| chr11_-_36619771 | 0.00 |
ENST00000311485.3 ENST00000527033.1 ENST00000532616.1 |
RAG2 |
recombination activating gene 2 |
| chr2_-_216003127 | 0.00 |
ENST00000412081.1 ENST00000272895.7 |
ABCA12 |
ATP-binding cassette, sub-family A (ABC1), member 12 |
| chr1_+_31883048 | 0.00 |
ENST00000536859.1 |
SERINC2 |
serine incorporator 2 |
| chr10_+_114710211 | 0.00 |
ENST00000349937.2 ENST00000369397.4 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr20_+_42187608 | 0.00 |
ENST00000373100.1 |
SGK2 |
serum/glucocorticoid regulated kinase 2 |
| chr3_+_77088989 | 0.00 |
ENST00000461745.1 |
ROBO2 |
roundabout, axon guidance receptor, homolog 2 (Drosophila) |
| chr6_-_133035185 | 0.00 |
ENST00000367928.4 |
VNN1 |
vanin 1 |
| chr3_+_111718173 | 0.00 |
ENST00000494932.1 |
TAGLN3 |
transgelin 3 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0061289 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.0 | 0.3 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.0 | 0.3 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.1 | GO:1904580 | regulation of intracellular mRNA localization(GO:1904580) positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.0 | 0.2 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.0 | 0.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 0.2 | GO:0010748 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 | 0.1 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.4 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.1 | GO:1904647 | response to rotenone(GO:1904647) |
| 0.0 | 0.2 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.1 | GO:0014740 | negative regulation of muscle hyperplasia(GO:0014740) |
| 0.0 | 0.2 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.1 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.0 | 0.1 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.1 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.0 | 0.4 | GO:0021670 | lateral ventricle development(GO:0021670) forebrain morphogenesis(GO:0048853) |
| 0.0 | 0.2 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.1 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.0 | 0.1 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.2 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.1 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.0 | 0.2 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.0 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.5 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.0 | 0.1 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.1 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.4 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.0 | 0.2 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.0 | GO:0031780 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.0 | 0.2 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |