Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
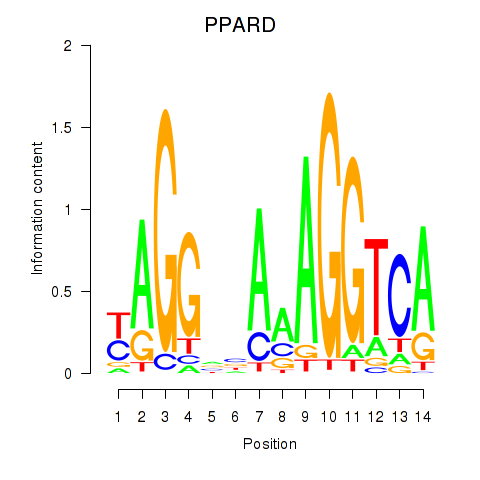
Results for PPARD
Z-value: 0.27
Transcription factors associated with PPARD
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PPARD
|
ENSG00000112033.9 | PPARD |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PPARD | hg19_v2_chr6_+_35310391_35310410 | 0.66 | 7.2e-02 | Click! |
Activity profile of PPARD motif
Sorted Z-values of PPARD motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PPARD
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_43885252 | 0.45 |
ENST00000453782.1 ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B |
creatine kinase, mitochondrial 1B |
| chr15_+_43985084 | 0.44 |
ENST00000434505.1 ENST00000411750.1 |
CKMT1A |
creatine kinase, mitochondrial 1A |
| chr19_-_10687907 | 0.32 |
ENST00000589348.1 |
AP1M2 |
adaptor-related protein complex 1, mu 2 subunit |
| chr19_-_10687948 | 0.31 |
ENST00000592285.1 |
AP1M2 |
adaptor-related protein complex 1, mu 2 subunit |
| chr19_-_10687983 | 0.28 |
ENST00000587069.1 |
AP1M2 |
adaptor-related protein complex 1, mu 2 subunit |
| chr17_+_48610074 | 0.20 |
ENST00000503690.1 ENST00000514874.1 ENST00000537145.1 ENST00000541226.1 |
EPN3 |
epsin 3 |
| chr2_+_206547215 | 0.19 |
ENST00000360409.3 ENST00000540178.1 ENST00000540841.1 ENST00000355117.4 ENST00000450507.1 ENST00000417189.1 |
NRP2 |
neuropilin 2 |
| chr12_-_53320245 | 0.17 |
ENST00000552150.1 |
KRT8 |
keratin 8 |
| chr2_+_95940186 | 0.13 |
ENST00000403131.2 ENST00000317668.4 ENST00000317620.9 |
PROM2 |
prominin 2 |
| chr1_+_62417957 | 0.12 |
ENST00000307297.7 ENST00000543708.1 |
INADL |
InaD-like (Drosophila) |
| chr11_-_119599794 | 0.12 |
ENST00000264025.3 |
PVRL1 |
poliovirus receptor-related 1 (herpesvirus entry mediator C) |
| chr5_-_16509101 | 0.11 |
ENST00000399793.2 |
FAM134B |
family with sequence similarity 134, member B |
| chr1_+_156123318 | 0.11 |
ENST00000368285.3 |
SEMA4A |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr4_+_140586922 | 0.10 |
ENST00000265498.1 ENST00000506797.1 |
MGST2 |
microsomal glutathione S-transferase 2 |
| chr17_+_37894179 | 0.09 |
ENST00000577695.1 ENST00000309156.4 ENST00000309185.3 |
GRB7 |
growth factor receptor-bound protein 7 |
| chr17_-_39728303 | 0.08 |
ENST00000588431.1 ENST00000246662.4 |
KRT9 |
keratin 9 |
| chr6_+_33172407 | 0.08 |
ENST00000374662.3 |
HSD17B8 |
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr16_-_4401258 | 0.07 |
ENST00000577031.1 |
PAM16 |
presequence translocase-associated motor 16 homolog (S. cerevisiae) |
| chr16_-_4401284 | 0.07 |
ENST00000318059.3 |
PAM16 |
presequence translocase-associated motor 16 homolog (S. cerevisiae) |
| chr7_+_112090483 | 0.07 |
ENST00000403825.3 ENST00000429071.1 |
IFRD1 |
interferon-related developmental regulator 1 |
| chr14_+_32546145 | 0.05 |
ENST00000556611.1 ENST00000539826.2 |
ARHGAP5 |
Rho GTPase activating protein 5 |
| chr14_+_32546274 | 0.05 |
ENST00000396582.2 |
ARHGAP5 |
Rho GTPase activating protein 5 |
| chr2_+_159651821 | 0.04 |
ENST00000309950.3 ENST00000409042.1 |
DAPL1 |
death associated protein-like 1 |
| chr14_-_51561784 | 0.04 |
ENST00000360392.4 |
TRIM9 |
tripartite motif containing 9 |
| chr2_-_99871570 | 0.04 |
ENST00000333017.2 ENST00000409679.1 ENST00000423306.1 |
LYG2 |
lysozyme G-like 2 |
| chr17_-_34270668 | 0.04 |
ENST00000293274.4 |
LYZL6 |
lysozyme-like 6 |
| chr17_-_34270697 | 0.04 |
ENST00000585556.1 |
LYZL6 |
lysozyme-like 6 |
| chrX_-_148571884 | 0.03 |
ENST00000537071.1 |
IDS |
iduronate 2-sulfatase |
| chr22_-_42526802 | 0.03 |
ENST00000359033.4 ENST00000389970.3 ENST00000360608.5 |
CYP2D6 |
cytochrome P450, family 2, subfamily D, polypeptide 6 |
| chr4_-_7069760 | 0.03 |
ENST00000264954.4 |
GRPEL1 |
GrpE-like 1, mitochondrial (E. coli) |
| chr12_-_95467267 | 0.03 |
ENST00000330677.7 |
NR2C1 |
nuclear receptor subfamily 2, group C, member 1 |
| chr11_-_61658853 | 0.03 |
ENST00000525588.1 ENST00000540820.1 |
FADS3 |
fatty acid desaturase 3 |
| chr22_-_29784519 | 0.03 |
ENST00000357586.2 ENST00000356015.2 ENST00000432560.2 ENST00000317368.7 |
AP1B1 |
adaptor-related protein complex 1, beta 1 subunit |
| chr6_-_33548006 | 0.03 |
ENST00000374467.3 |
BAK1 |
BCL2-antagonist/killer 1 |
| chr15_+_75494214 | 0.03 |
ENST00000394987.4 |
C15orf39 |
chromosome 15 open reading frame 39 |
| chr11_-_116968987 | 0.03 |
ENST00000434315.2 ENST00000292055.4 ENST00000375288.1 ENST00000542607.1 ENST00000445177.1 ENST00000375300.1 ENST00000446921.2 |
SIK3 |
SIK family kinase 3 |
| chr19_+_11546440 | 0.03 |
ENST00000589126.1 ENST00000588269.1 ENST00000587509.1 ENST00000592741.1 ENST00000593101.1 ENST00000587327.1 |
PRKCSH |
protein kinase C substrate 80K-H |
| chr19_+_11546153 | 0.03 |
ENST00000591946.1 ENST00000252455.2 ENST00000412601.1 |
PRKCSH |
protein kinase C substrate 80K-H |
| chr1_-_11107280 | 0.03 |
ENST00000400897.3 ENST00000400898.3 |
MASP2 |
mannan-binding lectin serine peptidase 2 |
| chr1_+_202317855 | 0.02 |
ENST00000356764.2 |
PPP1R12B |
protein phosphatase 1, regulatory subunit 12B |
| chr14_-_25045446 | 0.02 |
ENST00000216336.2 |
CTSG |
cathepsin G |
| chr6_-_33547975 | 0.02 |
ENST00000442998.2 ENST00000360661.5 |
BAK1 |
BCL2-antagonist/killer 1 |
| chr11_-_61659006 | 0.02 |
ENST00000278829.2 |
FADS3 |
fatty acid desaturase 3 |
| chr6_+_143999072 | 0.02 |
ENST00000440869.2 ENST00000367582.3 ENST00000451827.2 |
PHACTR2 |
phosphatase and actin regulator 2 |
| chr1_+_6511651 | 0.02 |
ENST00000434576.1 |
ESPN |
espin |
| chr6_+_30295036 | 0.02 |
ENST00000376659.5 ENST00000428555.1 |
TRIM39 |
tripartite motif containing 39 |
| chr19_+_10196981 | 0.02 |
ENST00000591813.1 |
C19orf66 |
chromosome 19 open reading frame 66 |
| chr15_-_82555000 | 0.02 |
ENST00000557844.1 ENST00000359445.3 ENST00000268206.7 |
EFTUD1 |
elongation factor Tu GTP binding domain containing 1 |
| chr11_-_45939374 | 0.02 |
ENST00000533151.1 ENST00000241041.3 |
PEX16 |
peroxisomal biogenesis factor 16 |
| chr19_+_11546093 | 0.02 |
ENST00000591462.1 |
PRKCSH |
protein kinase C substrate 80K-H |
| chr19_+_51273721 | 0.02 |
ENST00000270590.4 |
GPR32 |
G protein-coupled receptor 32 |
| chr7_-_87342564 | 0.02 |
ENST00000265724.3 ENST00000416177.1 |
ABCB1 |
ATP-binding cassette, sub-family B (MDR/TAP), member 1 |
| chr19_+_39616410 | 0.02 |
ENST00000602004.1 ENST00000599470.1 ENST00000321944.4 ENST00000593480.1 ENST00000358301.3 ENST00000593690.1 ENST00000599386.1 |
PAK4 |
p21 protein (Cdc42/Rac)-activated kinase 4 |
| chr6_+_32006042 | 0.02 |
ENST00000418967.2 |
CYP21A2 |
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr1_-_120311517 | 0.02 |
ENST00000369406.3 ENST00000544913.2 |
HMGCS2 |
3-hydroxy-3-methylglutaryl-CoA synthase 2 (mitochondrial) |
| chr5_+_34757309 | 0.02 |
ENST00000397449.1 |
RAI14 |
retinoic acid induced 14 |
| chr1_+_55107449 | 0.02 |
ENST00000421030.2 ENST00000545244.1 ENST00000339553.5 ENST00000409996.1 ENST00000454855.2 |
MROH7 |
maestro heat-like repeat family member 7 |
| chr6_+_32006159 | 0.01 |
ENST00000478281.1 ENST00000471671.1 ENST00000435122.2 |
CYP21A2 |
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr2_-_159313214 | 0.01 |
ENST00000409889.1 ENST00000283233.5 ENST00000536771.1 |
CCDC148 |
coiled-coil domain containing 148 |
| chr15_+_82555125 | 0.01 |
ENST00000566205.1 ENST00000339465.5 ENST00000569120.1 ENST00000566861.1 |
FAM154B |
family with sequence similarity 154, member B |
| chr21_+_27107672 | 0.01 |
ENST00000400075.3 |
GABPA |
GA binding protein transcription factor, alpha subunit 60kDa |
| chr12_-_109219937 | 0.01 |
ENST00000546697.1 |
SSH1 |
slingshot protein phosphatase 1 |
| chr6_+_142468383 | 0.01 |
ENST00000367621.1 ENST00000452973.2 |
VTA1 |
vesicle (multivesicular body) trafficking 1 |
| chr7_+_143174966 | 0.01 |
ENST00000408916.1 |
TAS2R41 |
taste receptor, type 2, member 41 |
| chr16_-_74808710 | 0.01 |
ENST00000219368.3 ENST00000544337.1 |
FA2H |
fatty acid 2-hydroxylase |
| chr2_+_219433588 | 0.01 |
ENST00000295701.5 |
RQCD1 |
RCD1 required for cell differentiation1 homolog (S. pombe) |
| chr11_+_64073699 | 0.01 |
ENST00000405666.1 ENST00000468670.1 |
ESRRA |
estrogen-related receptor alpha |
| chr6_+_43266063 | 0.01 |
ENST00000372574.3 |
SLC22A7 |
solute carrier family 22 (organic anion transporter), member 7 |
| chr2_-_207024134 | 0.01 |
ENST00000457011.1 ENST00000440274.1 ENST00000432169.1 |
NDUFS1 |
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr1_-_11863171 | 0.01 |
ENST00000376592.1 |
MTHFR |
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr6_+_142468361 | 0.01 |
ENST00000367630.4 |
VTA1 |
vesicle (multivesicular body) trafficking 1 |
| chr2_+_73441350 | 0.01 |
ENST00000389501.4 |
SMYD5 |
SMYD family member 5 |
| chr8_-_80942061 | 0.01 |
ENST00000519386.1 |
MRPS28 |
mitochondrial ribosomal protein S28 |
| chr8_-_80942467 | 0.01 |
ENST00000518271.1 ENST00000276585.4 ENST00000521605.1 |
MRPS28 |
mitochondrial ribosomal protein S28 |
| chr19_-_49339080 | 0.01 |
ENST00000595764.1 |
HSD17B14 |
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr1_-_197036364 | 0.01 |
ENST00000367412.1 |
F13B |
coagulation factor XIII, B polypeptide |
| chr16_-_75285380 | 0.00 |
ENST00000393420.6 ENST00000162330.5 |
BCAR1 |
breast cancer anti-estrogen resistance 1 |
| chr1_-_29508321 | 0.00 |
ENST00000546138.1 |
SRSF4 |
serine/arginine-rich splicing factor 4 |
| chr8_-_80942139 | 0.00 |
ENST00000521434.1 ENST00000519120.1 ENST00000520946.1 |
MRPS28 |
mitochondrial ribosomal protein S28 |
| chr5_+_14664762 | 0.00 |
ENST00000284274.4 |
FAM105B |
family with sequence similarity 105, member B |
| chr1_+_43855545 | 0.00 |
ENST00000372450.4 ENST00000310739.4 |
SZT2 |
seizure threshold 2 homolog (mouse) |
| chr1_+_202317815 | 0.00 |
ENST00000608999.1 ENST00000336894.4 ENST00000480184.1 |
PPP1R12B |
protein phosphatase 1, regulatory subunit 12B |
| chrX_+_2746850 | 0.00 |
ENST00000381163.3 ENST00000338623.5 ENST00000542787.1 |
GYG2 |
glycogenin 2 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.1 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.1 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.0 | 0.2 | GO:0097490 | sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 0.0 | 0.1 | GO:0048597 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 | 0.9 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.0 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.0 | 0.2 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.1 | GO:0044393 | microspike(GO:0044393) |
| 0.0 | 0.9 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.0 | 0.1 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |