Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
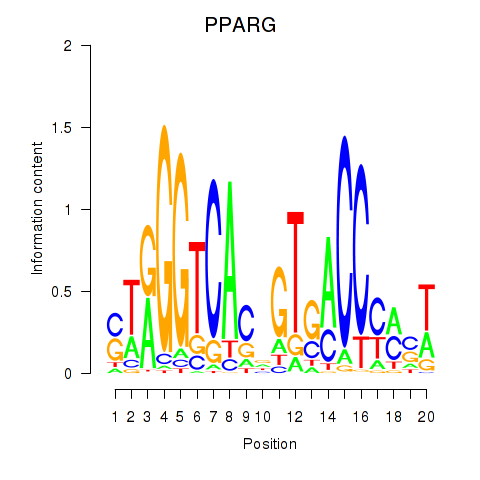
Results for PPARG
Z-value: 0.82
Transcription factors associated with PPARG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PPARG
|
ENSG00000132170.15 | PPARG |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PPARG | hg19_v2_chr3_+_12329358_12329393, hg19_v2_chr3_+_12330560_12330579, hg19_v2_chr3_+_12392971_12392983, hg19_v2_chr3_+_12329397_12329433 | 0.67 | 6.9e-02 | Click! |
Activity profile of PPARG motif
Sorted Z-values of PPARG motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PPARG
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_51587502 | 0.91 |
ENST00000156499.2 ENST00000391802.1 |
KLK14 |
kallikrein-related peptidase 14 |
| chr19_+_50016411 | 0.73 |
ENST00000426395.3 ENST00000600273.1 ENST00000599988.1 |
FCGRT |
Fc fragment of IgG, receptor, transporter, alpha |
| chr5_+_156696362 | 0.67 |
ENST00000377576.3 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
| chr15_+_33010175 | 0.65 |
ENST00000300177.4 ENST00000560677.1 ENST00000560830.1 |
GREM1 |
gremlin 1, DAN family BMP antagonist |
| chrX_-_51812268 | 0.56 |
ENST00000486010.1 ENST00000497164.1 ENST00000360134.6 ENST00000485287.1 ENST00000335504.5 ENST00000431659.1 |
MAGED4B |
melanoma antigen family D, 4B |
| chr19_+_50016610 | 0.53 |
ENST00000596975.1 |
FCGRT |
Fc fragment of IgG, receptor, transporter, alpha |
| chr3_+_99357319 | 0.52 |
ENST00000452013.1 ENST00000261037.3 ENST00000273342.4 |
COL8A1 |
collagen, type VIII, alpha 1 |
| chr3_-_145968923 | 0.49 |
ENST00000493382.1 ENST00000354952.2 ENST00000383083.2 |
PLSCR4 |
phospholipid scramblase 4 |
| chr16_+_8768422 | 0.48 |
ENST00000268251.8 ENST00000564714.1 |
ABAT |
4-aminobutyrate aminotransferase |
| chr8_+_70404996 | 0.46 |
ENST00000402687.4 ENST00000419716.3 |
SULF1 |
sulfatase 1 |
| chrX_+_51928002 | 0.44 |
ENST00000375626.3 |
MAGED4 |
melanoma antigen family D, 4 |
| chr3_-_145968857 | 0.43 |
ENST00000433593.2 ENST00000476202.1 ENST00000460885.1 |
PLSCR4 |
phospholipid scramblase 4 |
| chrX_+_9433048 | 0.43 |
ENST00000217964.7 |
TBL1X |
transducin (beta)-like 1X-linked |
| chr11_-_74022658 | 0.42 |
ENST00000427714.2 ENST00000331597.4 |
P4HA3 |
prolyl 4-hydroxylase, alpha polypeptide III |
| chr2_+_112895939 | 0.41 |
ENST00000331203.2 ENST00000409903.1 ENST00000409667.3 ENST00000409450.3 |
FBLN7 |
fibulin 7 |
| chr11_+_117070904 | 0.38 |
ENST00000529792.1 |
TAGLN |
transgelin |
| chr6_-_52441713 | 0.35 |
ENST00000182527.3 |
TRAM2 |
translocation associated membrane protein 2 |
| chr14_+_24630465 | 0.35 |
ENST00000557894.1 ENST00000559284.1 ENST00000560275.1 |
IRF9 |
interferon regulatory factor 9 |
| chr11_-_1771797 | 0.35 |
ENST00000340134.4 |
IFITM10 |
interferon induced transmembrane protein 10 |
| chr22_+_31277661 | 0.35 |
ENST00000454145.1 ENST00000453621.1 ENST00000431368.1 ENST00000535268.1 |
OSBP2 |
oxysterol binding protein 2 |
| chr3_+_69812701 | 0.33 |
ENST00000472437.1 |
MITF |
microphthalmia-associated transcription factor |
| chr3_+_69812877 | 0.33 |
ENST00000457080.1 ENST00000328528.6 |
MITF |
microphthalmia-associated transcription factor |
| chr22_-_24641027 | 0.32 |
ENST00000398292.3 ENST00000263112.7 ENST00000418439.2 ENST00000424217.1 ENST00000327365.4 |
GGT5 |
gamma-glutamyltransferase 5 |
| chr8_-_13134045 | 0.31 |
ENST00000512044.2 |
DLC1 |
deleted in liver cancer 1 |
| chr15_+_41056218 | 0.31 |
ENST00000260447.4 |
GCHFR |
GTP cyclohydrolase I feedback regulator |
| chr1_-_38218577 | 0.31 |
ENST00000540011.1 |
EPHA10 |
EPH receptor A10 |
| chr15_+_41056255 | 0.31 |
ENST00000561160.1 ENST00000559445.1 |
GCHFR |
GTP cyclohydrolase I feedback regulator |
| chr6_-_169364429 | 0.30 |
ENST00000444586.1 |
RP3-495K2.3 |
RP3-495K2.3 |
| chr17_+_48172639 | 0.30 |
ENST00000503176.1 ENST00000503614.1 |
PDK2 |
pyruvate dehydrogenase kinase, isozyme 2 |
| chr11_+_117070037 | 0.29 |
ENST00000392951.4 ENST00000525531.1 ENST00000278968.6 |
TAGLN |
transgelin |
| chr1_+_215256467 | 0.28 |
ENST00000391894.2 ENST00000444842.2 |
KCNK2 |
potassium channel, subfamily K, member 2 |
| chr2_+_11295624 | 0.28 |
ENST00000402361.1 ENST00000428481.1 |
PQLC3 |
PQ loop repeat containing 3 |
| chr4_-_1670632 | 0.27 |
ENST00000461064.1 |
FAM53A |
family with sequence similarity 53, member A |
| chr14_+_21152259 | 0.27 |
ENST00000555835.1 ENST00000336811.6 |
RNASE4 ANG |
ribonuclease, RNase A family, 4 angiogenin, ribonuclease, RNase A family, 5 |
| chr5_+_32710736 | 0.27 |
ENST00000415685.2 |
NPR3 |
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr13_+_107029084 | 0.27 |
ENST00000444865.1 |
LINC00460 |
long intergenic non-protein coding RNA 460 |
| chr19_+_35629702 | 0.26 |
ENST00000351325.4 |
FXYD1 |
FXYD domain containing ion transport regulator 1 |
| chr14_+_105212297 | 0.25 |
ENST00000556623.1 ENST00000555674.1 |
ADSSL1 |
adenylosuccinate synthase like 1 |
| chr19_+_35630022 | 0.25 |
ENST00000589209.1 |
FXYD1 |
FXYD domain containing ion transport regulator 1 |
| chr16_-_67969888 | 0.24 |
ENST00000574576.2 |
PSMB10 |
proteasome (prosome, macropain) subunit, beta type, 10 |
| chrX_-_106959631 | 0.24 |
ENST00000486554.1 ENST00000372390.4 |
TSC22D3 |
TSC22 domain family, member 3 |
| chr17_-_74236382 | 0.24 |
ENST00000592271.1 ENST00000319945.6 ENST00000269391.6 |
RNF157 |
ring finger protein 157 |
| chr19_-_50143452 | 0.23 |
ENST00000246792.3 |
RRAS |
related RAS viral (r-ras) oncogene homolog |
| chr9_+_140119618 | 0.23 |
ENST00000359069.2 |
C9orf169 |
chromosome 9 open reading frame 169 |
| chr1_+_201617264 | 0.23 |
ENST00000367296.4 |
NAV1 |
neuron navigator 1 |
| chr22_-_24322660 | 0.23 |
ENST00000404092.1 |
DDT |
D-dopachrome tautomerase |
| chr15_-_89089860 | 0.22 |
ENST00000558413.1 ENST00000564406.1 ENST00000268148.8 |
DET1 |
de-etiolated homolog 1 (Arabidopsis) |
| chrX_+_54947229 | 0.22 |
ENST00000442098.1 ENST00000430420.1 ENST00000453081.1 ENST00000173898.7 ENST00000319167.8 ENST00000375022.4 ENST00000399736.1 ENST00000440072.1 ENST00000420798.2 ENST00000431115.1 ENST00000440759.1 ENST00000375041.2 |
TRO |
trophinin |
| chr17_+_39975544 | 0.22 |
ENST00000544340.1 |
FKBP10 |
FK506 binding protein 10, 65 kDa |
| chr14_+_105953204 | 0.22 |
ENST00000409393.2 |
CRIP1 |
cysteine-rich protein 1 (intestinal) |
| chr14_+_100848311 | 0.22 |
ENST00000542471.2 |
WDR25 |
WD repeat domain 25 |
| chrX_+_153409678 | 0.21 |
ENST00000369951.4 |
OPN1LW |
opsin 1 (cone pigments), long-wave-sensitive |
| chr1_+_201617450 | 0.21 |
ENST00000295624.6 ENST00000367297.4 ENST00000367300.3 |
NAV1 |
neuron navigator 1 |
| chr11_-_64052111 | 0.21 |
ENST00000394532.3 ENST00000394531.3 ENST00000309032.3 |
BAD |
BCL2-associated agonist of cell death |
| chr14_+_21467414 | 0.21 |
ENST00000554422.1 ENST00000298681.4 |
SLC39A2 |
solute carrier family 39 (zinc transporter), member 2 |
| chr6_-_24911195 | 0.21 |
ENST00000259698.4 |
FAM65B |
family with sequence similarity 65, member B |
| chr14_+_105953246 | 0.21 |
ENST00000392531.3 |
CRIP1 |
cysteine-rich protein 1 (intestinal) |
| chr13_+_107028897 | 0.21 |
ENST00000439790.1 ENST00000435024.1 |
LINC00460 |
long intergenic non-protein coding RNA 460 |
| chr3_+_9958758 | 0.20 |
ENST00000383812.4 ENST00000438091.1 ENST00000295981.3 ENST00000436503.1 ENST00000403601.3 ENST00000416074.2 ENST00000455057.1 |
IL17RC |
interleukin 17 receptor C |
| chr1_-_95392635 | 0.20 |
ENST00000538964.1 ENST00000394202.4 ENST00000370206.4 |
CNN3 |
calponin 3, acidic |
| chrX_+_102862834 | 0.20 |
ENST00000372627.5 ENST00000243286.3 |
TCEAL3 |
transcription elongation factor A (SII)-like 3 |
| chr11_+_308143 | 0.20 |
ENST00000399817.4 |
IFITM2 |
interferon induced transmembrane protein 2 |
| chr20_+_49411523 | 0.20 |
ENST00000371608.2 |
BCAS4 |
breast carcinoma amplified sequence 4 |
| chr1_+_203274639 | 0.19 |
ENST00000290551.4 |
BTG2 |
BTG family, member 2 |
| chr11_+_2923619 | 0.19 |
ENST00000380574.1 |
SLC22A18 |
solute carrier family 22, member 18 |
| chr13_-_28024681 | 0.19 |
ENST00000381116.1 ENST00000381120.3 ENST00000431572.2 |
MTIF3 |
mitochondrial translational initiation factor 3 |
| chr19_+_56152262 | 0.19 |
ENST00000325333.5 ENST00000590190.1 |
ZNF580 |
zinc finger protein 580 |
| chr17_-_41984835 | 0.19 |
ENST00000520406.1 ENST00000518478.1 ENST00000522172.1 ENST00000461854.1 ENST00000521178.1 ENST00000520305.1 ENST00000523501.1 ENST00000520241.1 |
MPP2 |
membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2) |
| chr19_+_4343691 | 0.19 |
ENST00000597036.1 |
MPND |
MPN domain containing |
| chr2_-_231860596 | 0.18 |
ENST00000441063.1 ENST00000434094.1 ENST00000418330.1 ENST00000457803.1 ENST00000414876.1 ENST00000446741.1 ENST00000426904.1 |
AC105344.2 |
SPATA3 antisense RNA 1 (head to head) |
| chr5_-_70272055 | 0.18 |
ENST00000514857.2 |
NAIP |
NLR family, apoptosis inhibitory protein |
| chr19_-_10445399 | 0.18 |
ENST00000592945.1 |
ICAM3 |
intercellular adhesion molecule 3 |
| chr22_+_30792846 | 0.18 |
ENST00000312932.9 ENST00000428195.1 |
SEC14L2 |
SEC14-like 2 (S. cerevisiae) |
| chr2_+_231860830 | 0.18 |
ENST00000424440.1 ENST00000452881.1 ENST00000433428.2 ENST00000455816.1 ENST00000440792.1 ENST00000423134.1 |
SPATA3 |
spermatogenesis associated 3 |
| chr5_+_178977546 | 0.18 |
ENST00000319449.4 ENST00000377001.2 |
RUFY1 |
RUN and FYVE domain containing 1 |
| chr16_+_20912382 | 0.18 |
ENST00000396052.2 |
LYRM1 |
LYR motif containing 1 |
| chr6_+_83903061 | 0.18 |
ENST00000369724.4 ENST00000539997.1 |
RWDD2A |
RWD domain containing 2A |
| chr8_-_97247759 | 0.18 |
ENST00000518406.1 ENST00000523920.1 ENST00000287022.5 |
UQCRB |
ubiquinol-cytochrome c reductase binding protein |
| chr20_+_54933971 | 0.17 |
ENST00000371384.3 ENST00000437418.1 |
FAM210B |
family with sequence similarity 210, member B |
| chr12_+_113796347 | 0.17 |
ENST00000545182.2 ENST00000280800.3 |
PLBD2 |
phospholipase B domain containing 2 |
| chr11_+_2923499 | 0.17 |
ENST00000449793.2 |
SLC22A18 |
solute carrier family 22, member 18 |
| chr3_+_48507210 | 0.17 |
ENST00000433541.1 ENST00000422277.2 ENST00000436480.2 ENST00000444177.1 |
TREX1 |
three prime repair exonuclease 1 |
| chr1_-_111970353 | 0.17 |
ENST00000369732.3 |
OVGP1 |
oviductal glycoprotein 1, 120kDa |
| chr1_-_46642154 | 0.17 |
ENST00000540385.1 |
PIK3R3 |
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr19_+_18451391 | 0.17 |
ENST00000269919.6 ENST00000604499.2 ENST00000595066.1 ENST00000252813.5 |
PGPEP1 |
pyroglutamyl-peptidase I |
| chr20_+_31870927 | 0.16 |
ENST00000253354.1 |
BPIFB1 |
BPI fold containing family B, member 1 |
| chr19_+_52873166 | 0.16 |
ENST00000424032.2 ENST00000600321.1 ENST00000344085.5 ENST00000597976.1 ENST00000422689.2 |
ZNF880 |
zinc finger protein 880 |
| chr2_+_30369807 | 0.16 |
ENST00000379520.3 ENST00000379519.3 ENST00000261353.4 |
YPEL5 |
yippee-like 5 (Drosophila) |
| chr10_-_28623368 | 0.16 |
ENST00000441595.2 |
MPP7 |
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr12_+_133264156 | 0.16 |
ENST00000317479.3 ENST00000543589.1 |
PXMP2 |
peroxisomal membrane protein 2, 22kDa |
| chr3_+_48507621 | 0.16 |
ENST00000456089.1 |
TREX1 |
three prime repair exonuclease 1 |
| chr16_+_20912075 | 0.16 |
ENST00000219168.4 |
LYRM1 |
LYR motif containing 1 |
| chr19_-_9896672 | 0.16 |
ENST00000589412.1 ENST00000586814.1 |
ZNF846 |
zinc finger protein 846 |
| chr22_+_20008595 | 0.15 |
ENST00000398042.2 ENST00000450664.1 ENST00000327374.4 ENST00000432883.1 |
TANGO2 |
transport and golgi organization 2 homolog (Drosophila) |
| chr7_+_75932863 | 0.15 |
ENST00000429938.1 |
HSPB1 |
heat shock 27kDa protein 1 |
| chr12_-_120884175 | 0.15 |
ENST00000546954.1 |
TRIAP1 |
TP53 regulated inhibitor of apoptosis 1 |
| chr2_-_200322723 | 0.15 |
ENST00000417098.1 |
SATB2 |
SATB homeobox 2 |
| chr11_-_65793948 | 0.15 |
ENST00000312106.5 |
CATSPER1 |
cation channel, sperm associated 1 |
| chr9_-_35112376 | 0.15 |
ENST00000488109.2 |
FAM214B |
family with sequence similarity 214, member B |
| chr2_-_69664586 | 0.15 |
ENST00000303698.3 ENST00000394305.1 ENST00000410022.2 |
NFU1 |
NFU1 iron-sulfur cluster scaffold homolog (S. cerevisiae) |
| chr20_+_62152077 | 0.15 |
ENST00000370179.3 ENST00000370177.1 |
PPDPF |
pancreatic progenitor cell differentiation and proliferation factor |
| chr3_+_9958870 | 0.15 |
ENST00000413608.1 |
IL17RC |
interleukin 17 receptor C |
| chr2_+_201994042 | 0.14 |
ENST00000417748.1 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chr2_-_69664549 | 0.14 |
ENST00000450796.2 ENST00000484177.1 |
NFU1 |
NFU1 iron-sulfur cluster scaffold homolog (S. cerevisiae) |
| chr13_-_28024269 | 0.14 |
ENST00000405591.2 |
MTIF3 |
mitochondrial translational initiation factor 3 |
| chr16_+_30383613 | 0.14 |
ENST00000568749.1 |
MYLPF |
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr14_+_95047725 | 0.14 |
ENST00000554760.1 ENST00000554866.1 ENST00000329597.7 ENST00000556775.1 |
SERPINA5 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 |
| chr1_-_113247543 | 0.14 |
ENST00000414971.1 ENST00000534717.1 |
RHOC |
ras homolog family member C |
| chr8_-_27850141 | 0.14 |
ENST00000524352.1 |
SCARA5 |
scavenger receptor class A, member 5 (putative) |
| chr18_+_5238549 | 0.14 |
ENST00000580684.1 |
LINC00667 |
long intergenic non-protein coding RNA 667 |
| chr11_+_67171358 | 0.14 |
ENST00000526387.1 |
TBC1D10C |
TBC1 domain family, member 10C |
| chr17_-_56032602 | 0.14 |
ENST00000577840.1 |
CUEDC1 |
CUE domain containing 1 |
| chr19_+_850985 | 0.14 |
ENST00000590230.1 |
ELANE |
elastase, neutrophil expressed |
| chr11_+_67171548 | 0.13 |
ENST00000542590.1 |
TBC1D10C |
TBC1 domain family, member 10C |
| chr3_-_183735651 | 0.13 |
ENST00000427120.2 ENST00000392579.2 ENST00000382494.2 ENST00000446941.2 |
ABCC5 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr19_+_52800410 | 0.13 |
ENST00000595962.1 ENST00000598016.1 ENST00000334564.7 ENST00000490272.1 |
ZNF480 |
zinc finger protein 480 |
| chr11_+_67171391 | 0.13 |
ENST00000312390.5 |
TBC1D10C |
TBC1 domain family, member 10C |
| chr3_+_38537960 | 0.13 |
ENST00000453767.1 |
EXOG |
endo/exonuclease (5'-3'), endonuclease G-like |
| chr6_-_112575687 | 0.13 |
ENST00000521398.1 ENST00000424408.2 ENST00000243219.3 |
LAMA4 |
laminin, alpha 4 |
| chr9_+_17134980 | 0.13 |
ENST00000380647.3 |
CNTLN |
centlein, centrosomal protein |
| chr17_-_76899275 | 0.13 |
ENST00000322630.2 ENST00000586713.1 |
DDC8 |
Protein DDC8 homolog |
| chr10_-_48438974 | 0.13 |
ENST00000224605.2 |
GDF10 |
growth differentiation factor 10 |
| chr19_+_56717283 | 0.13 |
ENST00000376267.1 |
ZSCAN5C |
zinc finger and SCAN domain containing 5C |
| chr15_-_40331342 | 0.13 |
ENST00000559081.1 |
SRP14 |
signal recognition particle 14kDa (homologous Alu RNA binding protein) |
| chrX_+_135252050 | 0.13 |
ENST00000449474.1 ENST00000345434.3 |
FHL1 |
four and a half LIM domains 1 |
| chr19_+_4343584 | 0.13 |
ENST00000596722.1 |
MPND |
MPN domain containing |
| chr15_-_83316711 | 0.12 |
ENST00000568128.1 |
CPEB1 |
cytoplasmic polyadenylation element binding protein 1 |
| chr20_-_56286479 | 0.12 |
ENST00000265626.4 |
PMEPA1 |
prostate transmembrane protein, androgen induced 1 |
| chr16_+_67022476 | 0.12 |
ENST00000540947.2 |
CES4A |
carboxylesterase 4A |
| chr16_+_67022582 | 0.12 |
ENST00000541479.1 ENST00000338718.4 |
CES4A |
carboxylesterase 4A |
| chr16_+_67022633 | 0.12 |
ENST00000398354.1 ENST00000326686.5 |
CES4A |
carboxylesterase 4A |
| chr20_-_33999766 | 0.12 |
ENST00000349714.5 ENST00000438533.1 ENST00000359226.2 ENST00000374384.2 ENST00000374377.5 ENST00000407996.2 ENST00000424405.1 ENST00000542501.1 ENST00000397554.1 ENST00000540457.1 ENST00000374380.2 ENST00000374385.5 |
UQCC1 |
ubiquinol-cytochrome c reductase complex assembly factor 1 |
| chr7_+_150498610 | 0.12 |
ENST00000461345.1 |
TMEM176A |
transmembrane protein 176A |
| chr5_-_176936844 | 0.12 |
ENST00000510380.1 ENST00000510898.1 ENST00000357198.4 |
DOK3 |
docking protein 3 |
| chr16_+_476379 | 0.12 |
ENST00000434585.1 |
RAB11FIP3 |
RAB11 family interacting protein 3 (class II) |
| chr20_-_35402123 | 0.12 |
ENST00000373740.3 ENST00000426836.1 ENST00000373745.3 ENST00000448110.2 ENST00000438549.1 ENST00000447406.1 ENST00000373750.4 ENST00000373734.4 |
DSN1 |
DSN1, MIS12 kinetochore complex component |
| chr22_-_30162924 | 0.12 |
ENST00000344318.3 ENST00000397781.3 |
ZMAT5 |
zinc finger, matrin-type 5 |
| chrX_+_101380642 | 0.12 |
ENST00000372780.1 ENST00000329035.2 |
TCEAL2 |
transcription elongation factor A (SII)-like 2 |
| chr3_-_48594248 | 0.12 |
ENST00000545984.1 ENST00000232375.3 ENST00000416568.1 ENST00000383734.2 ENST00000541519.1 ENST00000412035.1 |
PFKFB4 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| chr17_-_79196799 | 0.12 |
ENST00000269392.4 |
AZI1 |
5-azacytidine induced 1 |
| chr16_+_57279248 | 0.12 |
ENST00000562023.1 ENST00000563234.1 |
ARL2BP |
ADP-ribosylation factor-like 2 binding protein |
| chr10_+_74870253 | 0.12 |
ENST00000544879.1 ENST00000537969.1 ENST00000372997.3 |
NUDT13 |
nudix (nucleoside diphosphate linked moiety X)-type motif 13 |
| chr10_+_4828815 | 0.12 |
ENST00000533295.1 |
AKR1E2 |
aldo-keto reductase family 1, member E2 |
| chr2_-_201374781 | 0.12 |
ENST00000359878.3 ENST00000409157.1 |
KCTD18 |
potassium channel tetramerization domain containing 18 |
| chr1_-_151762943 | 0.11 |
ENST00000368825.3 ENST00000368823.1 ENST00000458431.2 ENST00000368827.6 ENST00000368824.3 |
TDRKH |
tudor and KH domain containing |
| chr22_-_24096630 | 0.11 |
ENST00000248948.3 |
VPREB3 |
pre-B lymphocyte 3 |
| chr16_-_4850471 | 0.11 |
ENST00000592019.1 ENST00000586153.1 |
ROGDI |
rogdi homolog (Drosophila) |
| chr8_-_145115584 | 0.11 |
ENST00000426825.1 |
OPLAH |
5-oxoprolinase (ATP-hydrolysing) |
| chr3_+_195413160 | 0.11 |
ENST00000599448.1 |
LINC00969 |
long intergenic non-protein coding RNA 969 |
| chr12_-_56122761 | 0.11 |
ENST00000552164.1 ENST00000420846.3 ENST00000257857.4 |
CD63 |
CD63 molecule |
| chr19_+_507299 | 0.11 |
ENST00000359315.5 |
TPGS1 |
tubulin polyglutamylase complex subunit 1 |
| chr5_+_175976324 | 0.11 |
ENST00000261944.5 |
CDHR2 |
cadherin-related family member 2 |
| chr8_-_101118210 | 0.11 |
ENST00000360863.6 |
RGS22 |
regulator of G-protein signaling 22 |
| chr1_+_19923454 | 0.11 |
ENST00000602662.1 ENST00000602293.1 ENST00000322753.6 |
MINOS1-NBL1 MINOS1 |
MINOS1-NBL1 readthrough mitochondrial inner membrane organizing system 1 |
| chrX_+_153770421 | 0.11 |
ENST00000369609.5 ENST00000369607.1 |
IKBKG |
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr1_+_170501270 | 0.11 |
ENST00000367763.3 ENST00000367762.1 |
GORAB |
golgin, RAB6-interacting |
| chr18_-_5238525 | 0.11 |
ENST00000581170.1 ENST00000579933.1 ENST00000581067.1 |
RP11-835E18.5 LINC00526 |
RP11-835E18.5 long intergenic non-protein coding RNA 526 |
| chr11_+_59807748 | 0.11 |
ENST00000278855.2 ENST00000532905.1 |
PLAC1L |
oocyte secreted protein 2 |
| chr10_+_74870206 | 0.10 |
ENST00000357321.4 ENST00000349051.5 |
NUDT13 |
nudix (nucleoside diphosphate linked moiety X)-type motif 13 |
| chr15_+_44580955 | 0.10 |
ENST00000345795.2 ENST00000360824.3 |
CASC4 |
cancer susceptibility candidate 4 |
| chr5_+_78407602 | 0.10 |
ENST00000274353.5 ENST00000524080.1 |
BHMT |
betaine--homocysteine S-methyltransferase |
| chr11_-_65686586 | 0.10 |
ENST00000438576.2 |
C11orf68 |
chromosome 11 open reading frame 68 |
| chr4_-_186347099 | 0.10 |
ENST00000505357.1 ENST00000264689.6 |
UFSP2 |
UFM1-specific peptidase 2 |
| chr17_-_27044810 | 0.10 |
ENST00000395242.2 |
RAB34 |
RAB34, member RAS oncogene family |
| chr12_-_39299406 | 0.10 |
ENST00000331366.5 |
CPNE8 |
copine VIII |
| chr11_-_65686496 | 0.10 |
ENST00000449692.3 |
C11orf68 |
chromosome 11 open reading frame 68 |
| chr11_+_695787 | 0.10 |
ENST00000526170.1 ENST00000488769.1 |
TMEM80 |
transmembrane protein 80 |
| chr6_-_43478239 | 0.10 |
ENST00000372441.1 |
LRRC73 |
leucine rich repeat containing 73 |
| chr8_-_101118185 | 0.10 |
ENST00000523437.1 |
RGS22 |
regulator of G-protein signaling 22 |
| chr16_+_57279004 | 0.10 |
ENST00000219204.3 |
ARL2BP |
ADP-ribosylation factor-like 2 binding protein |
| chr4_+_41992489 | 0.10 |
ENST00000264451.7 |
SLC30A9 |
solute carrier family 30 (zinc transporter), member 9 |
| chr12_-_56123444 | 0.10 |
ENST00000546457.1 ENST00000549117.1 |
CD63 |
CD63 molecule |
| chr11_-_67210930 | 0.10 |
ENST00000453768.2 ENST00000545016.1 ENST00000341356.5 |
CORO1B |
coronin, actin binding protein, 1B |
| chr1_+_156096336 | 0.10 |
ENST00000504687.1 ENST00000473598.2 |
LMNA |
lamin A/C |
| chr7_+_150498783 | 0.10 |
ENST00000475536.1 ENST00000468689.1 |
TMEM176A |
transmembrane protein 176A |
| chr20_+_44563267 | 0.10 |
ENST00000372409.3 |
PCIF1 |
PDX1 C-terminal inhibiting factor 1 |
| chr17_+_48911942 | 0.09 |
ENST00000426127.1 |
WFIKKN2 |
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 |
| chr19_-_22018966 | 0.09 |
ENST00000599906.1 ENST00000354959.4 |
ZNF43 |
zinc finger protein 43 |
| chr12_-_39300071 | 0.09 |
ENST00000550863.1 |
CPNE8 |
copine VIII |
| chr1_-_47407111 | 0.09 |
ENST00000371904.4 |
CYP4A11 |
cytochrome P450, family 4, subfamily A, polypeptide 11 |
| chr9_+_101984577 | 0.09 |
ENST00000223641.4 |
SEC61B |
Sec61 beta subunit |
| chr8_-_101117847 | 0.09 |
ENST00000523287.1 ENST00000519092.1 |
RGS22 |
regulator of G-protein signaling 22 |
| chr6_+_33378738 | 0.09 |
ENST00000374512.3 ENST00000374516.3 |
PHF1 |
PHD finger protein 1 |
| chr19_-_47349395 | 0.09 |
ENST00000597020.1 |
AP2S1 |
adaptor-related protein complex 2, sigma 1 subunit |
| chr19_-_46405861 | 0.09 |
ENST00000322217.5 |
MYPOP |
Myb-related transcription factor, partner of profilin |
| chr22_+_30792980 | 0.09 |
ENST00000403484.1 ENST00000405717.3 ENST00000402592.3 |
SEC14L2 |
SEC14-like 2 (S. cerevisiae) |
| chr15_+_57998923 | 0.09 |
ENST00000380557.4 |
POLR2M |
polymerase (RNA) II (DNA directed) polypeptide M |
| chr6_+_30594619 | 0.09 |
ENST00000318999.7 ENST00000376485.4 ENST00000376478.2 ENST00000319027.5 ENST00000376483.4 ENST00000329992.8 ENST00000330083.5 |
ATAT1 |
alpha tubulin acetyltransferase 1 |
| chr15_-_40331356 | 0.09 |
ENST00000560773.1 ENST00000267884.6 |
SRP14 |
signal recognition particle 14kDa (homologous Alu RNA binding protein) |
| chr15_+_63414760 | 0.09 |
ENST00000557972.1 |
LACTB |
lactamase, beta |
| chr6_+_139094657 | 0.09 |
ENST00000332797.6 |
CCDC28A |
coiled-coil domain containing 28A |
| chr15_+_57998821 | 0.09 |
ENST00000299638.3 |
POLR2M |
polymerase (RNA) II (DNA directed) polypeptide M |
| chr18_+_52495426 | 0.09 |
ENST00000262094.5 |
RAB27B |
RAB27B, member RAS oncogene family |
| chr1_-_1009683 | 0.09 |
ENST00000453464.2 |
RNF223 |
ring finger protein 223 |
| chr3_-_157251383 | 0.09 |
ENST00000487753.1 ENST00000489602.1 ENST00000461299.1 ENST00000479987.1 |
VEPH1 |
ventricular zone expressed PH domain-containing 1 |
| chr4_+_141178440 | 0.09 |
ENST00000394205.3 |
SCOC |
short coiled-coil protein |
| chr2_-_3521518 | 0.09 |
ENST00000382093.5 |
ADI1 |
acireductone dioxygenase 1 |
| chr17_+_45286706 | 0.09 |
ENST00000393450.1 ENST00000572303.1 |
MYL4 |
myosin, light chain 4, alkali; atrial, embryonic |
| chr1_+_23037323 | 0.09 |
ENST00000544305.1 ENST00000374630.3 ENST00000400191.3 ENST00000374632.3 |
EPHB2 |
EPH receptor B2 |
| chr19_-_46285646 | 0.09 |
ENST00000458663.2 |
DMPK |
dystrophia myotonica-protein kinase |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:1900158 | negative regulation of osteoclast proliferation(GO:0090291) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.2 | 0.6 | GO:0043105 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.1 | 1.3 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.1 | 0.5 | GO:1904448 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.1 | 0.5 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 0.3 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.1 | 0.4 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.1 | 0.5 | GO:0010732 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.1 | 0.3 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.1 | 0.2 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.1 | 0.2 | GO:0060139 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.1 | 0.7 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 0.9 | GO:0007320 | insemination(GO:0007320) |
| 0.1 | 0.2 | GO:0051685 | maintenance of ER location(GO:0051685) |
| 0.1 | 0.4 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 0.3 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 0.1 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.0 | 0.9 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.3 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.3 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.0 | 0.2 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.0 | 0.4 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.1 | GO:0090675 | intermicrovillar adhesion(GO:0090675) |
| 0.0 | 0.1 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 0.0 | 0.1 | GO:0019520 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.0 | 0.3 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.5 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.1 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.0 | 0.4 | GO:0097396 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.0 | 0.2 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.3 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.0 | 0.1 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.2 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.0 | 0.1 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.1 | GO:1903238 | positive regulation of leukocyte tethering or rolling(GO:1903238) |
| 0.0 | 0.1 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.2 | GO:1902019 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.1 | GO:0055073 | cadmium ion homeostasis(GO:0055073) divalent metal ion export(GO:0070839) |
| 0.0 | 0.4 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.1 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.0 | 0.1 | GO:0070221 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.0 | 0.2 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.3 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.1 | GO:0052250 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.0 | 0.4 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.1 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.2 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.1 | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.2 | GO:0038033 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) |
| 0.0 | 0.0 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.0 | 0.0 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.0 | 0.0 | GO:2000397 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.1 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.0 | 0.2 | GO:2000680 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.0 | 0.0 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.0 | 0.2 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.1 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.0 | 0.3 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.1 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.1 | GO:0045144 | meiotic sister chromatid segregation(GO:0045144) |
| 0.0 | 0.3 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.2 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.1 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.1 | GO:0060373 | T-tubule organization(GO:0033292) detection of muscle stretch(GO:0035995) regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.3 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.1 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.7 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.0 | GO:0070781 | response to biotin(GO:0070781) |
| 0.0 | 0.1 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.2 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 | 0.3 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.0 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 0.0 | 0.2 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 0.1 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.1 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.2 | GO:0042438 | melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.1 | 0.3 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 0.5 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.1 | 0.3 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.1 | GO:0036024 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.0 | 0.3 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.1 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) condensed chromosome inner kinetochore(GO:0000939) |
| 0.0 | 0.3 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.2 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.1 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.3 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.0 | 0.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.3 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.0 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.0 | 0.4 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.2 | 0.6 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.1 | 0.5 | GO:0032145 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.1 | 0.3 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.1 | 0.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.3 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.3 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.1 | 0.7 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 0.4 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.1 | 0.4 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.1 | 0.2 | GO:0033981 | D-dopachrome decarboxylase activity(GO:0033981) |
| 0.1 | 0.3 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.1 | 0.9 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.2 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.3 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.4 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.3 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.2 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.0 | 0.1 | GO:0050571 | 1,5-anhydro-D-fructose reductase activity(GO:0050571) |
| 0.0 | 0.3 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.1 | GO:0047150 | betaine-homocysteine S-methyltransferase activity(GO:0047150) |
| 0.0 | 0.1 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.0 | 0.1 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.2 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.2 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.1 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.0 | 0.6 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.1 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.0 | 0.1 | GO:0051139 | metal ion:proton antiporter activity(GO:0051139) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.1 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.2 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.4 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.3 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.1 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.3 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.0 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.0 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.0 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.7 | GO:0070888 | E-box binding(GO:0070888) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.8 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.4 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.4 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |