Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for PRDM1
Z-value: 0.80
Transcription factors associated with PRDM1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PRDM1
|
ENSG00000057657.10 | PRDM1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PRDM1 | hg19_v2_chr6_+_106546808_106546833 | -0.93 | 9.0e-04 | Click! |
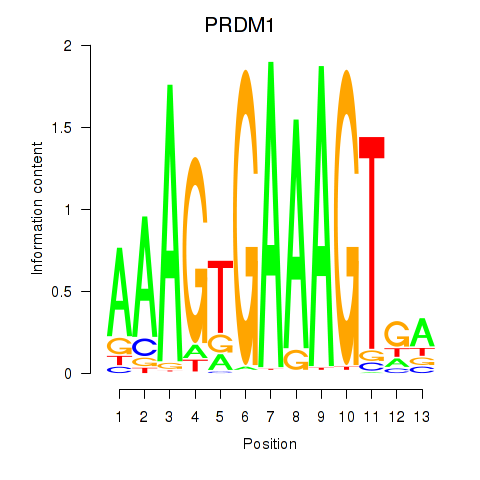
Activity profile of PRDM1 motif
Sorted Z-values of PRDM1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PRDM1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_92919043 | 1.69 |
ENST00000327111.3 |
NR2F1 |
nuclear receptor subfamily 2, group F, member 1 |
| chr8_+_97506033 | 1.65 |
ENST00000518385.1 |
SDC2 |
syndecan 2 |
| chr13_+_102104980 | 1.60 |
ENST00000545560.2 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr3_+_8543393 | 0.84 |
ENST00000157600.3 ENST00000415597.1 ENST00000535732.1 |
LMCD1 |
LIM and cysteine-rich domains 1 |
| chr6_-_167275991 | 0.83 |
ENST00000510118.1 |
RPS6KA2 |
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr6_-_167276033 | 0.80 |
ENST00000503859.1 ENST00000506565.1 |
RPS6KA2 |
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr3_+_69915385 | 0.76 |
ENST00000314589.5 |
MITF |
microphthalmia-associated transcription factor |
| chr17_+_41158742 | 0.75 |
ENST00000415816.2 ENST00000438323.2 |
IFI35 |
interferon-induced protein 35 |
| chrX_-_118827333 | 0.66 |
ENST00000360156.7 ENST00000354228.4 ENST00000489216.1 ENST00000354416.3 ENST00000394610.1 ENST00000343984.5 |
SEPT6 |
septin 6 |
| chr9_+_18474098 | 0.64 |
ENST00000327883.7 ENST00000431052.2 ENST00000380570.4 |
ADAMTSL1 |
ADAMTS-like 1 |
| chr3_+_8543561 | 0.63 |
ENST00000397386.3 |
LMCD1 |
LIM and cysteine-rich domains 1 |
| chr2_-_145275228 | 0.60 |
ENST00000427902.1 ENST00000409487.3 ENST00000470879.1 ENST00000435831.1 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr7_+_95401851 | 0.60 |
ENST00000447467.2 |
DYNC1I1 |
dynein, cytoplasmic 1, intermediate chain 1 |
| chr4_-_100242549 | 0.59 |
ENST00000305046.8 ENST00000394887.3 |
ADH1B |
alcohol dehydrogenase 1B (class I), beta polypeptide |
| chr3_-_49851313 | 0.57 |
ENST00000333486.3 |
UBA7 |
ubiquitin-like modifier activating enzyme 7 |
| chr8_+_77593448 | 0.57 |
ENST00000521891.2 |
ZFHX4 |
zinc finger homeobox 4 |
| chr15_-_37393406 | 0.56 |
ENST00000338564.5 ENST00000558313.1 ENST00000340545.5 |
MEIS2 |
Meis homeobox 2 |
| chr8_+_77593474 | 0.54 |
ENST00000455469.2 ENST00000050961.6 |
ZFHX4 |
zinc finger homeobox 4 |
| chr16_-_67970990 | 0.54 |
ENST00000358514.4 |
PSMB10 |
proteasome (prosome, macropain) subunit, beta type, 10 |
| chr3_+_8543533 | 0.53 |
ENST00000454244.1 |
LMCD1 |
LIM and cysteine-rich domains 1 |
| chr9_+_18474204 | 0.53 |
ENST00000276935.6 |
ADAMTSL1 |
ADAMTS-like 1 |
| chrX_-_63005405 | 0.50 |
ENST00000374878.1 ENST00000437457.2 |
ARHGEF9 |
Cdc42 guanine nucleotide exchange factor (GEF) 9 |
| chrX_-_118826784 | 0.45 |
ENST00000394616.4 |
SEPT6 |
septin 6 |
| chr4_+_142557717 | 0.45 |
ENST00000320650.4 ENST00000296545.7 |
IL15 |
interleukin 15 |
| chrX_-_118827113 | 0.44 |
ENST00000394617.2 |
SEPT6 |
septin 6 |
| chr5_+_112227311 | 0.43 |
ENST00000391338.1 |
ZRSR1 |
zinc finger (CCCH type), RNA-binding motif and serine/arginine rich 1 |
| chr4_+_142557771 | 0.42 |
ENST00000514653.1 |
IL15 |
interleukin 15 |
| chr8_-_116681221 | 0.41 |
ENST00000395715.3 |
TRPS1 |
trichorhinophalangeal syndrome I |
| chr11_-_96076334 | 0.39 |
ENST00000524717.1 |
MAML2 |
mastermind-like 2 (Drosophila) |
| chr3_-_114343039 | 0.38 |
ENST00000481632.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr5_-_93447333 | 0.38 |
ENST00000395965.3 ENST00000505869.1 ENST00000509163.1 |
FAM172A |
family with sequence similarity 172, member A |
| chr4_+_142558078 | 0.38 |
ENST00000529613.1 |
IL15 |
interleukin 15 |
| chr10_-_50747064 | 0.38 |
ENST00000355832.5 ENST00000603152.1 ENST00000447839.2 |
ERCC6 PGBD3 ERCC6-PGBD3 |
excision repair cross-complementing rodent repair deficiency, complementation group 6 piggyBac transposable element derived 3 ERCC6-PGBD3 readthrough |
| chr1_-_182361327 | 0.36 |
ENST00000331872.6 ENST00000311223.5 |
GLUL |
glutamate-ammonia ligase |
| chrX_+_10124977 | 0.36 |
ENST00000380833.4 |
CLCN4 |
chloride channel, voltage-sensitive 4 |
| chr2_+_231191875 | 0.36 |
ENST00000444636.1 ENST00000415673.2 ENST00000243810.6 ENST00000396563.4 |
SP140L |
SP140 nuclear body protein-like |
| chr19_+_49977818 | 0.34 |
ENST00000594009.1 ENST00000595510.1 |
FLT3LG |
fms-related tyrosine kinase 3 ligand |
| chr14_+_24605389 | 0.30 |
ENST00000382708.3 ENST00000561435.1 |
PSME1 |
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr1_-_182360918 | 0.30 |
ENST00000339526.4 |
GLUL |
glutamate-ammonia ligase |
| chr7_+_95401877 | 0.29 |
ENST00000524053.1 ENST00000324972.6 ENST00000537881.1 ENST00000437599.1 ENST00000359388.4 ENST00000413338.1 |
DYNC1I1 |
dynein, cytoplasmic 1, intermediate chain 1 |
| chr12_+_94542459 | 0.29 |
ENST00000258526.4 |
PLXNC1 |
plexin C1 |
| chr6_-_112575687 | 0.29 |
ENST00000521398.1 ENST00000424408.2 ENST00000243219.3 |
LAMA4 |
laminin, alpha 4 |
| chr12_+_94656297 | 0.28 |
ENST00000545312.1 |
PLXNC1 |
plexin C1 |
| chr5_+_49962495 | 0.27 |
ENST00000515175.1 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
| chr2_+_175260514 | 0.27 |
ENST00000424069.1 ENST00000427038.1 |
SCRN3 |
secernin 3 |
| chr6_-_137113604 | 0.26 |
ENST00000359015.4 |
MAP3K5 |
mitogen-activated protein kinase kinase kinase 5 |
| chr7_+_7811992 | 0.26 |
ENST00000406829.1 |
RPA3-AS1 |
RPA3 antisense RNA 1 |
| chr16_+_50775971 | 0.26 |
ENST00000311559.9 ENST00000564326.1 ENST00000566206.1 |
CYLD |
cylindromatosis (turban tumor syndrome) |
| chr2_+_175260451 | 0.25 |
ENST00000458563.1 ENST00000409673.3 ENST00000272732.6 ENST00000435964.1 |
SCRN3 |
secernin 3 |
| chr8_-_116680833 | 0.25 |
ENST00000220888.5 |
TRPS1 |
trichorhinophalangeal syndrome I |
| chr12_+_51318513 | 0.25 |
ENST00000332160.4 |
METTL7A |
methyltransferase like 7A |
| chr17_+_46908350 | 0.23 |
ENST00000258947.3 ENST00000509507.1 ENST00000448105.2 ENST00000570513.1 ENST00000509415.1 ENST00000513119.1 ENST00000416445.2 ENST00000508679.1 ENST00000505071.1 |
CALCOCO2 |
calcium binding and coiled-coil domain 2 |
| chr16_+_50775948 | 0.23 |
ENST00000569681.1 ENST00000569418.1 ENST00000540145.1 |
CYLD |
cylindromatosis (turban tumor syndrome) |
| chr12_-_54653313 | 0.23 |
ENST00000550411.1 ENST00000439541.2 |
CBX5 |
chromobox homolog 5 |
| chr21_+_17909594 | 0.23 |
ENST00000441820.1 ENST00000602280.1 |
LINC00478 |
long intergenic non-protein coding RNA 478 |
| chr2_+_166150541 | 0.23 |
ENST00000283256.6 |
SCN2A |
sodium channel, voltage-gated, type II, alpha subunit |
| chr2_-_231084617 | 0.22 |
ENST00000409815.2 |
SP110 |
SP110 nuclear body protein |
| chr1_+_110453109 | 0.22 |
ENST00000525659.1 |
CSF1 |
colony stimulating factor 1 (macrophage) |
| chr4_+_186990298 | 0.22 |
ENST00000296795.3 ENST00000513189.1 |
TLR3 |
toll-like receptor 3 |
| chr22_-_36635684 | 0.22 |
ENST00000358502.5 |
APOL2 |
apolipoprotein L, 2 |
| chr8_-_29120580 | 0.20 |
ENST00000524189.1 |
KIF13B |
kinesin family member 13B |
| chr3_+_185046676 | 0.20 |
ENST00000428617.1 ENST00000443863.1 |
MAP3K13 |
mitogen-activated protein kinase kinase kinase 13 |
| chr20_+_388791 | 0.20 |
ENST00000441733.1 ENST00000353660.3 |
RBCK1 |
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr4_-_54457783 | 0.20 |
ENST00000263925.7 ENST00000512247.1 |
LNX1 |
ligand of numb-protein X 1, E3 ubiquitin protein ligase |
| chr20_-_57089934 | 0.20 |
ENST00000439429.1 ENST00000371149.3 |
APCDD1L |
adenomatosis polyposis coli down-regulated 1-like |
| chr13_+_31309645 | 0.19 |
ENST00000380490.3 |
ALOX5AP |
arachidonate 5-lipoxygenase-activating protein |
| chr6_+_31540056 | 0.19 |
ENST00000418386.2 |
LTA |
lymphotoxin alpha |
| chr11_+_71710648 | 0.19 |
ENST00000260049.5 |
IL18BP |
interleukin 18 binding protein |
| chr14_+_24605361 | 0.19 |
ENST00000206451.6 ENST00000559123.1 |
PSME1 |
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr9_+_137533615 | 0.19 |
ENST00000371817.3 |
COL5A1 |
collagen, type V, alpha 1 |
| chr6_-_154831779 | 0.19 |
ENST00000607772.1 |
CNKSR3 |
CNKSR family member 3 |
| chr7_+_134464376 | 0.18 |
ENST00000454108.1 ENST00000361675.2 |
CALD1 |
caldesmon 1 |
| chr22_-_36556821 | 0.18 |
ENST00000531095.1 ENST00000397293.2 ENST00000349314.2 |
APOL3 |
apolipoprotein L, 3 |
| chr14_-_24615805 | 0.18 |
ENST00000560410.1 |
PSME2 |
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr20_+_30946106 | 0.18 |
ENST00000375687.4 ENST00000542461.1 |
ASXL1 |
additional sex combs like 1 (Drosophila) |
| chr6_+_32821924 | 0.17 |
ENST00000374859.2 ENST00000453265.2 |
PSMB9 |
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr11_+_71710973 | 0.17 |
ENST00000393707.4 |
IL18BP |
interleukin 18 binding protein |
| chr8_+_134203273 | 0.17 |
ENST00000250160.6 |
WISP1 |
WNT1 inducible signaling pathway protein 1 |
| chr10_-_82049424 | 0.17 |
ENST00000372213.3 |
MAT1A |
methionine adenosyltransferase I, alpha |
| chr14_-_24615523 | 0.17 |
ENST00000559056.1 |
PSME2 |
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr13_+_24144509 | 0.17 |
ENST00000248484.4 |
TNFRSF19 |
tumor necrosis factor receptor superfamily, member 19 |
| chr17_+_6659153 | 0.17 |
ENST00000441631.1 ENST00000438512.1 ENST00000346752.4 ENST00000361842.3 |
XAF1 |
XIAP associated factor 1 |
| chr6_+_32811885 | 0.16 |
ENST00000458296.1 ENST00000413039.1 ENST00000429600.1 ENST00000412095.1 ENST00000415067.1 ENST00000395330.1 |
TAPSAR1 PSMB9 |
TAP1 and PSMB8 antisense RNA 1 proteasome (prosome, macropain) subunit, beta type, 9 |
| chr13_+_108921977 | 0.16 |
ENST00000430559.1 ENST00000375887.4 |
TNFSF13B |
tumor necrosis factor (ligand) superfamily, member 13b |
| chr2_+_231280954 | 0.16 |
ENST00000409824.1 ENST00000409341.1 ENST00000409112.1 ENST00000340126.4 ENST00000341950.4 |
SP100 |
SP100 nuclear antigen |
| chr17_-_26694979 | 0.16 |
ENST00000438614.1 |
VTN |
vitronectin |
| chr22_-_31688381 | 0.16 |
ENST00000487265.2 |
PIK3IP1 |
phosphoinositide-3-kinase interacting protein 1 |
| chr1_-_205325850 | 0.16 |
ENST00000537168.1 |
KLHDC8A |
kelch domain containing 8A |
| chr5_-_149380698 | 0.15 |
ENST00000296736.3 |
TIGD6 |
tigger transposable element derived 6 |
| chr6_+_32811861 | 0.15 |
ENST00000453426.1 |
TAPSAR1 |
TAP1 and PSMB8 antisense RNA 1 |
| chr17_-_26695013 | 0.15 |
ENST00000555059.2 |
CTB-96E2.2 |
Homeobox protein SEBOX |
| chr8_-_54436491 | 0.15 |
ENST00000426023.1 |
RP11-400K9.4 |
RP11-400K9.4 |
| chr14_-_69263043 | 0.14 |
ENST00000408913.2 |
ZFP36L1 |
ZFP36 ring finger protein-like 1 |
| chr6_+_57182400 | 0.14 |
ENST00000607273.1 |
PRIM2 |
primase, DNA, polypeptide 2 (58kDa) |
| chr1_+_110453462 | 0.14 |
ENST00000488198.1 |
CSF1 |
colony stimulating factor 1 (macrophage) |
| chr16_-_31085514 | 0.14 |
ENST00000300849.4 |
ZNF668 |
zinc finger protein 668 |
| chr12_+_6561190 | 0.14 |
ENST00000544021.1 ENST00000266556.7 |
TAPBPL |
TAP binding protein-like |
| chr10_-_35104185 | 0.13 |
ENST00000374789.3 ENST00000374788.3 ENST00000346874.4 ENST00000374794.3 ENST00000350537.4 ENST00000374790.3 ENST00000374776.1 ENST00000374773.1 ENST00000545693.1 ENST00000545260.1 ENST00000340077.5 |
PARD3 |
par-3 family cell polarity regulator |
| chr7_+_134464414 | 0.13 |
ENST00000361901.2 |
CALD1 |
caldesmon 1 |
| chr3_+_119013185 | 0.13 |
ENST00000264245.4 |
ARHGAP31 |
Rho GTPase activating protein 31 |
| chr11_+_71709938 | 0.13 |
ENST00000393705.4 ENST00000337131.5 ENST00000531053.1 ENST00000404792.1 |
IL18BP |
interleukin 18 binding protein |
| chr15_-_37392086 | 0.13 |
ENST00000561208.1 |
MEIS2 |
Meis homeobox 2 |
| chr9_+_18474163 | 0.12 |
ENST00000380566.4 |
ADAMTSL1 |
ADAMTS-like 1 |
| chr2_-_231084820 | 0.12 |
ENST00000258382.5 ENST00000338556.3 |
SP110 |
SP110 nuclear body protein |
| chr2_+_69240302 | 0.12 |
ENST00000303714.4 |
ANTXR1 |
anthrax toxin receptor 1 |
| chr6_-_105584560 | 0.12 |
ENST00000336775.5 |
BVES |
blood vessel epicardial substance |
| chr11_+_46402297 | 0.12 |
ENST00000405308.2 |
MDK |
midkine (neurite growth-promoting factor 2) |
| chr13_+_24144796 | 0.11 |
ENST00000403372.2 |
TNFRSF19 |
tumor necrosis factor receptor superfamily, member 19 |
| chr18_+_18943554 | 0.11 |
ENST00000580732.2 |
GREB1L |
growth regulation by estrogen in breast cancer-like |
| chr6_-_32811771 | 0.10 |
ENST00000395339.3 ENST00000374882.3 |
PSMB8 |
proteasome (prosome, macropain) subunit, beta type, 8 |
| chr6_-_112575758 | 0.10 |
ENST00000431543.2 ENST00000453937.2 ENST00000368638.4 ENST00000389463.4 |
LAMA4 |
laminin, alpha 4 |
| chr2_+_239756671 | 0.09 |
ENST00000448943.2 |
TWIST2 |
twist family bHLH transcription factor 2 |
| chr10_-_112678692 | 0.09 |
ENST00000605742.1 |
BBIP1 |
BBSome interacting protein 1 |
| chr2_-_231084659 | 0.09 |
ENST00000258381.6 ENST00000358662.4 ENST00000455674.1 ENST00000392048.3 |
SP110 |
SP110 nuclear body protein |
| chr12_-_95941987 | 0.09 |
ENST00000537435.2 |
USP44 |
ubiquitin specific peptidase 44 |
| chr15_-_56209306 | 0.09 |
ENST00000506154.1 ENST00000338963.2 ENST00000508342.1 |
NEDD4 |
neural precursor cell expressed, developmentally down-regulated 4, E3 ubiquitin protein ligase |
| chr6_-_112575838 | 0.09 |
ENST00000455073.1 |
LAMA4 |
laminin, alpha 4 |
| chr8_+_77318769 | 0.09 |
ENST00000518732.1 |
RP11-706J10.1 |
long intergenic non-protein coding RNA 1111 |
| chr6_-_28220002 | 0.09 |
ENST00000377294.2 |
ZKSCAN4 |
zinc finger with KRAB and SCAN domains 4 |
| chr11_+_46402744 | 0.08 |
ENST00000533952.1 |
MDK |
midkine (neurite growth-promoting factor 2) |
| chr19_-_6670128 | 0.08 |
ENST00000245912.3 |
TNFSF14 |
tumor necrosis factor (ligand) superfamily, member 14 |
| chr16_+_50776021 | 0.07 |
ENST00000566679.2 ENST00000564634.1 ENST00000398568.2 |
CYLD |
cylindromatosis (turban tumor syndrome) |
| chr11_+_46402583 | 0.07 |
ENST00000359803.3 |
MDK |
midkine (neurite growth-promoting factor 2) |
| chr22_+_41697520 | 0.07 |
ENST00000352645.4 |
ZC3H7B |
zinc finger CCCH-type containing 7B |
| chr21_-_35899113 | 0.07 |
ENST00000492600.1 ENST00000481448.1 ENST00000381132.2 |
RCAN1 |
regulator of calcineurin 1 |
| chr22_-_31688431 | 0.07 |
ENST00000402249.3 ENST00000443175.1 ENST00000215912.5 ENST00000441972.1 |
PIK3IP1 |
phosphoinositide-3-kinase interacting protein 1 |
| chr15_+_71185148 | 0.07 |
ENST00000443425.2 ENST00000560755.1 |
LRRC49 |
leucine rich repeat containing 49 |
| chr2_-_70475586 | 0.07 |
ENST00000416149.2 |
TIA1 |
TIA1 cytotoxic granule-associated RNA binding protein |
| chr5_+_76506706 | 0.07 |
ENST00000340978.3 ENST00000346042.3 ENST00000264917.5 ENST00000342343.4 ENST00000333194.4 |
PDE8B |
phosphodiesterase 8B |
| chr2_-_175260368 | 0.06 |
ENST00000342016.3 ENST00000362053.5 |
CIR1 |
corepressor interacting with RBPJ, 1 |
| chr20_+_13976015 | 0.06 |
ENST00000217246.4 |
MACROD2 |
MACRO domain containing 2 |
| chrX_-_119694538 | 0.06 |
ENST00000371322.5 |
CUL4B |
cullin 4B |
| chr8_+_96145974 | 0.05 |
ENST00000315367.3 |
PLEKHF2 |
pleckstrin homology domain containing, family F (with FYVE domain) member 2 |
| chr2_+_231280908 | 0.05 |
ENST00000427101.2 ENST00000432979.1 |
SP100 |
SP100 nuclear antigen |
| chr6_-_112575912 | 0.05 |
ENST00000522006.1 ENST00000230538.7 ENST00000519932.1 |
LAMA4 |
laminin, alpha 4 |
| chr1_+_28099683 | 0.05 |
ENST00000373943.4 |
STX12 |
syntaxin 12 |
| chr10_+_22610124 | 0.04 |
ENST00000376663.3 |
BMI1 |
BMI1 polycomb ring finger oncogene |
| chr7_+_128355439 | 0.04 |
ENST00000315184.5 |
FAM71F1 |
family with sequence similarity 71, member F1 |
| chr4_-_74088800 | 0.04 |
ENST00000509867.2 |
ANKRD17 |
ankyrin repeat domain 17 |
| chr15_+_71184931 | 0.04 |
ENST00000560369.1 ENST00000260382.5 |
LRRC49 |
leucine rich repeat containing 49 |
| chr12_-_118498958 | 0.04 |
ENST00000315436.3 |
WSB2 |
WD repeat and SOCS box containing 2 |
| chr1_+_151372010 | 0.04 |
ENST00000290541.6 |
PSMB4 |
proteasome (prosome, macropain) subunit, beta type, 4 |
| chr22_-_30642782 | 0.04 |
ENST00000249075.3 |
LIF |
leukemia inhibitory factor |
| chr16_+_30675654 | 0.04 |
ENST00000287468.5 ENST00000395073.2 |
FBRS |
fibrosin |
| chr15_+_78832747 | 0.04 |
ENST00000560217.1 ENST00000044462.7 ENST00000559082.1 ENST00000559948.1 ENST00000413382.2 ENST00000559146.1 ENST00000558281.1 |
PSMA4 |
proteasome (prosome, macropain) subunit, alpha type, 4 |
| chr11_+_46402482 | 0.04 |
ENST00000441869.1 |
MDK |
midkine (neurite growth-promoting factor 2) |
| chr1_+_181003067 | 0.04 |
ENST00000434571.2 ENST00000367579.3 ENST00000282990.6 ENST00000367580.5 |
MR1 |
major histocompatibility complex, class I-related |
| chr3_-_149375783 | 0.04 |
ENST00000467467.1 ENST00000460517.1 ENST00000360632.3 |
WWTR1 |
WW domain containing transcription regulator 1 |
| chr6_-_33282024 | 0.03 |
ENST00000475304.1 ENST00000489157.1 |
TAPBP |
TAP binding protein (tapasin) |
| chr5_-_96143796 | 0.03 |
ENST00000296754.3 |
ERAP1 |
endoplasmic reticulum aminopeptidase 1 |
| chr12_-_52800139 | 0.03 |
ENST00000257974.2 |
KRT82 |
keratin 82 |
| chr10_-_21661870 | 0.03 |
ENST00000433460.1 |
RP11-275N1.1 |
RP11-275N1.1 |
| chr2_+_69240415 | 0.03 |
ENST00000409829.3 |
ANTXR1 |
anthrax toxin receptor 1 |
| chrX_-_133930285 | 0.03 |
ENST00000486347.1 ENST00000343004.5 |
FAM122B |
family with sequence similarity 122B |
| chr1_+_110453203 | 0.03 |
ENST00000357302.4 ENST00000344188.5 ENST00000329608.6 |
CSF1 |
colony stimulating factor 1 (macrophage) |
| chr19_+_24009879 | 0.03 |
ENST00000354585.4 |
RPSAP58 |
ribosomal protein SA pseudogene 58 |
| chr6_+_118869452 | 0.03 |
ENST00000357525.5 |
PLN |
phospholamban |
| chr6_+_26156551 | 0.03 |
ENST00000304218.3 |
HIST1H1E |
histone cluster 1, H1e |
| chr8_-_56986768 | 0.02 |
ENST00000523936.1 |
RPS20 |
ribosomal protein S20 |
| chr5_+_17444119 | 0.02 |
ENST00000503267.1 ENST00000504998.1 |
RP11-321E2.4 |
RP11-321E2.4 |
| chr6_+_146348810 | 0.02 |
ENST00000492807.2 |
GRM1 |
glutamate receptor, metabotropic 1 |
| chr4_-_100140331 | 0.02 |
ENST00000407820.2 ENST00000394897.1 ENST00000508558.1 ENST00000394899.2 |
ADH6 |
alcohol dehydrogenase 6 (class V) |
| chr2_-_202508169 | 0.02 |
ENST00000409883.2 |
TMEM237 |
transmembrane protein 237 |
| chr3_-_44552094 | 0.02 |
ENST00000436261.1 |
ZNF852 |
zinc finger protein 852 |
| chr6_+_108616243 | 0.02 |
ENST00000421954.1 |
LACE1 |
lactation elevated 1 |
| chr17_+_3379284 | 0.02 |
ENST00000263080.2 |
ASPA |
aspartoacylase |
| chr12_-_50561075 | 0.01 |
ENST00000422340.2 ENST00000317551.6 |
CERS5 |
ceramide synthase 5 |
| chr9_-_69229650 | 0.01 |
ENST00000416428.1 |
CBWD6 |
COBW domain containing 6 |
| chr14_-_23479331 | 0.01 |
ENST00000397377.1 ENST00000397379.3 ENST00000406429.2 ENST00000341470.4 ENST00000555998.1 ENST00000397376.2 ENST00000553675.1 ENST00000553931.1 ENST00000555575.1 ENST00000553958.1 ENST00000555098.1 ENST00000556419.1 ENST00000553606.1 ENST00000299088.6 ENST00000554179.1 ENST00000397382.4 |
C14orf93 |
chromosome 14 open reading frame 93 |
| chr1_+_199996702 | 0.01 |
ENST00000367362.3 |
NR5A2 |
nuclear receptor subfamily 5, group A, member 2 |
| chr11_-_40315640 | 0.01 |
ENST00000278198.2 |
LRRC4C |
leucine rich repeat containing 4C |
| chr6_-_33281979 | 0.01 |
ENST00000426633.2 ENST00000467025.1 |
TAPBP |
TAP binding protein (tapasin) |
| chr20_+_388679 | 0.01 |
ENST00000356286.5 ENST00000475269.1 |
RBCK1 |
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr6_-_32806483 | 0.01 |
ENST00000374899.4 |
TAP2 |
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr18_-_47017956 | 0.01 |
ENST00000584895.1 ENST00000332968.6 ENST00000580210.1 ENST00000579408.1 |
RPL17-C18orf32 RPL17 |
RPL17-C18orf32 readthrough ribosomal protein L17 |
| chr8_-_9009079 | 0.00 |
ENST00000519699.1 |
PPP1R3B |
protein phosphatase 1, regulatory subunit 3B |
| chr8_+_134203303 | 0.00 |
ENST00000519433.1 ENST00000517423.1 ENST00000377863.2 ENST00000220856.6 |
WISP1 |
WNT1 inducible signaling pathway protein 1 |
| chr10_+_18429671 | 0.00 |
ENST00000282343.8 |
CACNB2 |
calcium channel, voltage-dependent, beta 2 subunit |
| chr16_+_31085714 | 0.00 |
ENST00000300850.5 ENST00000564189.1 ENST00000428260.1 |
ZNF646 |
zinc finger protein 646 |
| chr6_-_132910877 | 0.00 |
ENST00000258034.2 |
TAAR5 |
trace amine associated receptor 5 |
| chr4_+_152020715 | 0.00 |
ENST00000274065.4 |
RPS3A |
ribosomal protein S3A |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 0.3 | 1.6 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.2 | 0.7 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.2 | 0.6 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.1 | 0.3 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.1 | 0.6 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 1.3 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.1 | 2.0 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 0.9 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.3 | GO:0060611 | mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) positive regulation of microglial cell migration(GO:1904141) |
| 0.1 | 0.2 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.1 | 0.2 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.2 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.1 | 0.6 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.2 | GO:0034344 | microglial cell activation involved in immune response(GO:0002282) type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.1 | 0.2 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.1 | 0.3 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.1 | GO:1904580 | regulation of intracellular mRNA localization(GO:1904580) positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.0 | 0.1 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.0 | 0.2 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.1 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) |
| 0.0 | 0.4 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.4 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.2 | GO:0031296 | positive regulation of germinal center formation(GO:0002636) B cell costimulation(GO:0031296) |
| 0.0 | 0.6 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.2 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.0 | GO:0086092 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) |
| 0.0 | 0.2 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:0044111 | development involved in symbiotic interaction(GO:0044111) |
| 0.0 | 0.2 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 0.2 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.0 | 0.5 | GO:1900115 | extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 1.9 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.0 | 0.7 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.2 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.6 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.0 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.3 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.0 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0032173 | septin ring(GO:0005940) septin collar(GO:0032173) |
| 0.1 | 0.8 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 1.0 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.4 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 0.3 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.1 | 0.3 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.2 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.7 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.0 | 0.9 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.4 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.5 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.6 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 1.6 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.3 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.2 | GO:0036020 | endolysosome membrane(GO:0036020) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.2 | 1.7 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 0.5 | GO:0042007 | interleukin-18 binding(GO:0042007) |
| 0.1 | 0.8 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.6 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 0.6 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.1 | 0.2 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 0.2 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.1 | 0.4 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.6 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.3 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.9 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 1.1 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.6 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 1.6 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.5 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.2 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 1.8 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.6 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.4 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.6 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.6 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 1.2 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.6 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 1.6 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.4 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 1.9 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.2 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 1.1 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 1.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.1 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.9 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |