Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for PRDM14
Z-value: 0.67
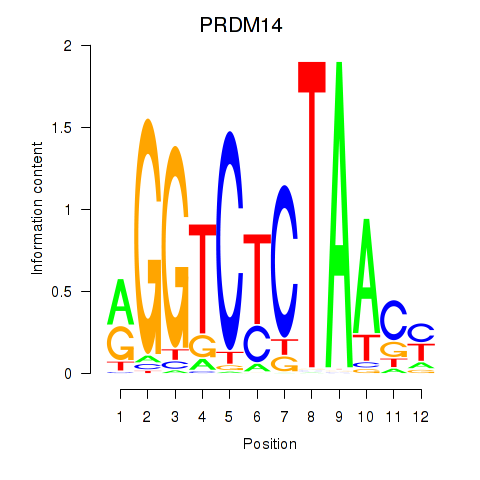
Transcription factors associated with PRDM14
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PRDM14
|
ENSG00000147596.3 | PRDM14 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PRDM14 | hg19_v2_chr8_-_70983506_70983562 | 0.07 | 8.7e-01 | Click! |
Activity profile of PRDM14 motif
Sorted Z-values of PRDM14 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PRDM14
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_19651654 | 1.10 |
ENST00000395555.3 |
ALDH3A1 |
aldehyde dehydrogenase 3 family, member A1 |
| chr17_-_19651668 | 1.01 |
ENST00000494157.2 ENST00000225740.6 |
ALDH3A1 |
aldehyde dehydrogenase 3 family, member A1 |
| chr11_+_15136462 | 0.97 |
ENST00000379556.3 ENST00000424273.1 |
INSC |
inscuteable homolog (Drosophila) |
| chr2_-_145278475 | 0.93 |
ENST00000558170.2 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr5_+_32711419 | 0.63 |
ENST00000265074.8 |
NPR3 |
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr1_-_31845914 | 0.62 |
ENST00000373713.2 |
FABP3 |
fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) |
| chr6_+_72926145 | 0.55 |
ENST00000425662.2 ENST00000453976.2 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr3_-_114477962 | 0.50 |
ENST00000471418.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr5_+_32710736 | 0.45 |
ENST00000415685.2 |
NPR3 |
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr11_+_64879317 | 0.36 |
ENST00000526809.1 ENST00000279263.7 ENST00000524986.1 ENST00000534371.1 ENST00000540748.1 ENST00000525385.1 ENST00000345348.5 ENST00000531321.1 ENST00000529414.1 ENST00000526085.1 ENST00000530750.1 |
TM7SF2 |
transmembrane 7 superfamily member 2 |
| chr21_+_26934165 | 0.36 |
ENST00000456917.1 |
MIR155HG |
MIR155 host gene (non-protein coding) |
| chr2_-_145277882 | 0.35 |
ENST00000465070.1 ENST00000444559.1 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr1_-_182642017 | 0.34 |
ENST00000367557.4 ENST00000258302.4 |
RGS8 |
regulator of G-protein signaling 8 |
| chr11_-_105892937 | 0.30 |
ENST00000301919.4 ENST00000534458.1 ENST00000530108.1 ENST00000530788.1 |
MSANTD4 |
Myb/SANT-like DNA-binding domain containing 4 with coiled-coils |
| chr2_-_192711968 | 0.29 |
ENST00000304141.4 |
SDPR |
serum deprivation response |
| chrX_+_51636629 | 0.28 |
ENST00000375722.1 ENST00000326587.7 ENST00000375695.2 |
MAGED1 |
melanoma antigen family D, 1 |
| chr4_+_81187753 | 0.28 |
ENST00000312465.7 ENST00000456523.3 |
FGF5 |
fibroblast growth factor 5 |
| chr19_-_53757941 | 0.27 |
ENST00000594517.1 ENST00000601413.1 ENST00000594681.1 |
ZNF677 |
zinc finger protein 677 |
| chr1_+_65730385 | 0.26 |
ENST00000263441.7 ENST00000395325.3 |
DNAJC6 |
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr17_+_36873677 | 0.25 |
ENST00000471200.1 |
MLLT6 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6 |
| chr1_+_109102652 | 0.24 |
ENST00000370035.3 ENST00000405454.1 |
FAM102B |
family with sequence similarity 102, member B |
| chr12_+_72332618 | 0.22 |
ENST00000333850.3 |
TPH2 |
tryptophan hydroxylase 2 |
| chrX_+_119292467 | 0.22 |
ENST00000371388.3 |
RHOXF2 |
Rhox homeobox family, member 2 |
| chr20_-_3154162 | 0.21 |
ENST00000360342.3 |
LZTS3 |
Homo sapiens leucine zipper, putative tumor suppressor family member 3 (LZTS3), mRNA. |
| chr8_+_110346546 | 0.19 |
ENST00000521662.1 ENST00000521688.1 ENST00000520147.1 |
ENY2 |
enhancer of yellow 2 homolog (Drosophila) |
| chr6_-_109777128 | 0.18 |
ENST00000358807.3 ENST00000358577.3 |
MICAL1 |
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr17_+_75450075 | 0.18 |
ENST00000592951.1 |
SEPT9 |
septin 9 |
| chr5_-_135528822 | 0.16 |
ENST00000607574.1 |
AC009014.3 |
AC009014.3 |
| chr3_+_16926441 | 0.16 |
ENST00000418129.2 ENST00000396755.2 |
PLCL2 |
phospholipase C-like 2 |
| chr19_-_49314169 | 0.15 |
ENST00000597011.1 ENST00000601681.1 |
BCAT2 |
branched chain amino-acid transaminase 2, mitochondrial |
| chr19_-_49314269 | 0.14 |
ENST00000545387.2 ENST00000316273.6 ENST00000402551.1 ENST00000598162.1 ENST00000599246.1 |
BCAT2 |
branched chain amino-acid transaminase 2, mitochondrial |
| chr2_+_101437487 | 0.14 |
ENST00000427413.1 ENST00000542504.1 |
NPAS2 |
neuronal PAS domain protein 2 |
| chr4_+_4861385 | 0.14 |
ENST00000382723.4 |
MSX1 |
msh homeobox 1 |
| chr20_-_48770174 | 0.13 |
ENST00000341698.2 |
TMEM189-UBE2V1 |
TMEM189-UBE2V1 readthrough |
| chr19_+_3572758 | 0.13 |
ENST00000416526.1 |
HMG20B |
high mobility group 20B |
| chr11_-_62474803 | 0.11 |
ENST00000533982.1 ENST00000360796.5 |
BSCL2 |
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr1_+_27114418 | 0.11 |
ENST00000078527.4 |
PIGV |
phosphatidylinositol glycan anchor biosynthesis, class V |
| chr1_-_42384343 | 0.10 |
ENST00000372584.1 |
HIVEP3 |
human immunodeficiency virus type I enhancer binding protein 3 |
| chr1_+_156052354 | 0.10 |
ENST00000368301.2 |
LMNA |
lamin A/C |
| chr11_-_65625014 | 0.09 |
ENST00000534784.1 |
CFL1 |
cofilin 1 (non-muscle) |
| chr12_+_49760639 | 0.09 |
ENST00000549538.1 ENST00000548654.1 ENST00000550643.1 ENST00000548710.1 ENST00000549179.1 ENST00000548377.1 |
SPATS2 |
spermatogenesis associated, serine-rich 2 |
| chr1_+_27114589 | 0.08 |
ENST00000431541.1 ENST00000449950.2 ENST00000374145.1 |
PIGV |
phosphatidylinositol glycan anchor biosynthesis, class V |
| chr12_-_49351228 | 0.07 |
ENST00000541959.1 ENST00000447318.2 |
ARF3 |
ADP-ribosylation factor 3 |
| chr2_-_228244013 | 0.06 |
ENST00000304568.3 |
TM4SF20 |
transmembrane 4 L six family member 20 |
| chr12_-_49351148 | 0.05 |
ENST00000398092.4 ENST00000539611.1 |
RP11-302B13.5 ARF3 |
ADP-ribosylation factor 3 ADP-ribosylation factor 3 |
| chr3_+_37035263 | 0.05 |
ENST00000458205.2 ENST00000539477.1 |
MLH1 |
mutL homolog 1 |
| chr16_-_55866997 | 0.04 |
ENST00000360526.3 ENST00000361503.4 |
CES1 |
carboxylesterase 1 |
| chr3_+_10289707 | 0.04 |
ENST00000287652.4 |
TATDN2 |
TatD DNase domain containing 2 |
| chr3_+_155838337 | 0.03 |
ENST00000490337.1 ENST00000389636.5 |
KCNAB1 |
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr3_+_37035289 | 0.03 |
ENST00000455445.2 ENST00000441265.1 ENST00000435176.1 ENST00000429117.1 ENST00000536378.1 |
MLH1 |
mutL homolog 1 |
| chr17_+_41924511 | 0.03 |
ENST00000377203.4 ENST00000539718.1 ENST00000588884.1 ENST00000293396.8 ENST00000586233.1 |
CD300LG |
CD300 molecule-like family member g |
| chrX_+_41193407 | 0.03 |
ENST00000457138.2 ENST00000441189.2 |
DDX3X |
DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked |
| chr11_-_65624415 | 0.03 |
ENST00000524553.1 ENST00000527344.1 |
CFL1 |
cofilin 1 (non-muscle) |
| chr7_+_150211918 | 0.02 |
ENST00000313543.4 |
GIMAP7 |
GTPase, IMAP family member 7 |
| chr1_+_50571949 | 0.02 |
ENST00000357083.4 |
ELAVL4 |
ELAV like neuron-specific RNA binding protein 4 |
| chr12_-_76742183 | 0.02 |
ENST00000393262.3 |
BBS10 |
Bardet-Biedl syndrome 10 |
| chr19_-_6279932 | 0.02 |
ENST00000252674.7 |
MLLT1 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 1 |
| chr17_-_46691990 | 0.02 |
ENST00000576562.1 |
HOXB8 |
homeobox B8 |
| chr3_-_155394152 | 0.01 |
ENST00000494598.1 |
PLCH1 |
phospholipase C, eta 1 |
| chr17_-_73775839 | 0.01 |
ENST00000592643.1 ENST00000591890.1 ENST00000587171.1 ENST00000254810.4 ENST00000589599.1 |
H3F3B |
H3 histone, family 3B (H3.3B) |
| chr19_+_39109735 | 0.01 |
ENST00000593149.1 ENST00000248342.4 ENST00000538434.1 ENST00000588934.1 ENST00000545173.2 ENST00000589307.1 ENST00000586513.1 ENST00000591409.1 ENST00000592558.1 |
EIF3K |
eukaryotic translation initiation factor 3, subunit K |
| chr6_-_46048116 | 0.00 |
ENST00000185206.6 |
CLIC5 |
chloride intracellular channel 5 |
| chr12_+_121647962 | 0.00 |
ENST00000542067.1 |
P2RX4 |
purinergic receptor P2X, ligand-gated ion channel, 4 |
| chr15_+_69850521 | 0.00 |
ENST00000558781.1 |
RP11-279F6.1 |
RP11-279F6.1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.2 | 0.6 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.1 | 0.3 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.1 | 1.1 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.1 | 0.2 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.1 | 0.2 | GO:0042427 | serotonin biosynthetic process(GO:0042427) |
| 0.1 | 0.2 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.1 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.0 | 0.5 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.1 | GO:0043060 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 0.0 | 0.3 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 2.1 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 0.1 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.1 | GO:2000769 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.3 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.2 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.3 | GO:0023019 | signal transduction involved in regulation of gene expression(GO:0023019) |
| 0.0 | 0.0 | GO:0090119 | vesicle-mediated cholesterol transport(GO:0090119) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 0.1 | GO:0005715 | late recombination nodule(GO:0005715) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.2 | 0.6 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.2 | 1.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.4 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.3 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.2 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.0 | 0.2 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 1.3 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) R-SMAD binding(GO:0070412) |
| 0.0 | 0.1 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |