Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
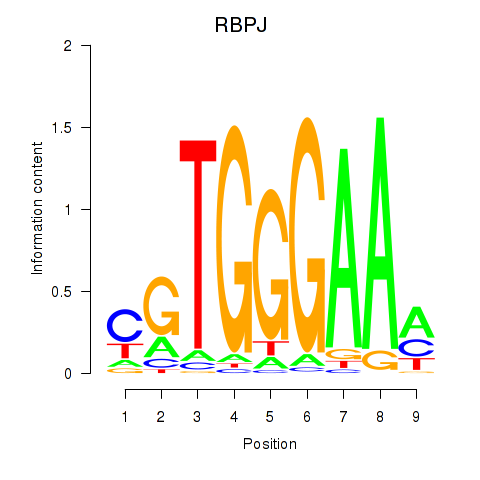
Results for RBPJ
Z-value: 0.42
Transcription factors associated with RBPJ
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RBPJ
|
ENSG00000168214.16 | RBPJ |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RBPJ | hg19_v2_chr4_+_26322185_26322334 | 0.76 | 2.9e-02 | Click! |
Activity profile of RBPJ motif
Sorted Z-values of RBPJ motif
Network of associatons between targets according to the STRING database.
First level regulatory network of RBPJ
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_100712352 | 0.72 |
ENST00000471714.1 ENST00000284322.5 |
ABI3BP |
ABI family, member 3 (NESH) binding protein |
| chr10_+_31608054 | 0.51 |
ENST00000320985.10 ENST00000361642.5 ENST00000560721.2 ENST00000558440.1 ENST00000424869.1 ENST00000542815.3 |
ZEB1 |
zinc finger E-box binding homeobox 1 |
| chr1_-_935491 | 0.29 |
ENST00000304952.6 |
HES4 |
hes family bHLH transcription factor 4 |
| chr3_+_69812877 | 0.28 |
ENST00000457080.1 ENST00000328528.6 |
MITF |
microphthalmia-associated transcription factor |
| chr20_+_43211149 | 0.26 |
ENST00000372886.1 |
PKIG |
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr5_-_124080203 | 0.22 |
ENST00000504926.1 |
ZNF608 |
zinc finger protein 608 |
| chr1_-_935361 | 0.21 |
ENST00000484667.2 |
HES4 |
hes family bHLH transcription factor 4 |
| chr2_-_79315112 | 0.19 |
ENST00000305089.3 |
REG1B |
regenerating islet-derived 1 beta |
| chr1_-_85870177 | 0.18 |
ENST00000542148.1 |
DDAH1 |
dimethylarginine dimethylaminohydrolase 1 |
| chr14_-_53417732 | 0.16 |
ENST00000399304.3 ENST00000395631.2 ENST00000341590.3 ENST00000343279.4 |
FERMT2 |
fermitin family member 2 |
| chr10_-_38146510 | 0.16 |
ENST00000395867.3 |
ZNF248 |
zinc finger protein 248 |
| chr3_+_69812701 | 0.15 |
ENST00000472437.1 |
MITF |
microphthalmia-associated transcription factor |
| chr2_-_111230393 | 0.15 |
ENST00000447537.2 ENST00000413601.2 |
LIMS3L LIMS3L |
LIM and senescent cell antigen-like-containing domain protein 3; Uncharacterized protein; cDNA FLJ59124, highly similar to Particularly interesting newCys-His protein; cDNA, FLJ79109, highly similar to Particularly interesting newCys-His protein LIM and senescent cell antigen-like domains 3-like |
| chr10_-_4285923 | 0.14 |
ENST00000418372.1 ENST00000608792.1 |
LINC00702 |
long intergenic non-protein coding RNA 702 |
| chr5_-_139943830 | 0.14 |
ENST00000412920.3 ENST00000511201.2 ENST00000356738.2 ENST00000354402.5 ENST00000358580.5 ENST00000508496.2 |
APBB3 |
amyloid beta (A4) precursor protein-binding, family B, member 3 |
| chr10_+_118305435 | 0.14 |
ENST00000369221.2 |
PNLIP |
pancreatic lipase |
| chr15_+_74218787 | 0.13 |
ENST00000261921.7 |
LOXL1 |
lysyl oxidase-like 1 |
| chr17_+_2699697 | 0.13 |
ENST00000254695.8 ENST00000366401.4 ENST00000542807.1 |
RAP1GAP2 |
RAP1 GTPase activating protein 2 |
| chr1_-_935519 | 0.13 |
ENST00000428771.2 |
HES4 |
hes family bHLH transcription factor 4 |
| chr2_+_110656005 | 0.13 |
ENST00000437679.2 |
LIMS3 |
LIM and senescent cell antigen-like domains 3 |
| chr11_+_118478313 | 0.12 |
ENST00000356063.5 |
PHLDB1 |
pleckstrin homology-like domain, family B, member 1 |
| chr7_+_90338712 | 0.12 |
ENST00000265741.3 ENST00000406263.1 |
CDK14 |
cyclin-dependent kinase 14 |
| chr10_-_44144292 | 0.12 |
ENST00000374433.2 |
ZNF32 |
zinc finger protein 32 |
| chr1_-_146644036 | 0.12 |
ENST00000425272.2 |
PRKAB2 |
protein kinase, AMP-activated, beta 2 non-catalytic subunit |
| chr2_+_109271481 | 0.12 |
ENST00000542845.1 ENST00000393314.2 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr10_-_44144152 | 0.12 |
ENST00000395797.1 |
ZNF32 |
zinc finger protein 32 |
| chr19_-_39694894 | 0.11 |
ENST00000318438.6 |
SYCN |
syncollin |
| chr5_+_140792614 | 0.11 |
ENST00000398610.2 |
PCDHGA10 |
protocadherin gamma subfamily A, 10 |
| chr10_+_115439630 | 0.11 |
ENST00000369318.3 |
CASP7 |
caspase 7, apoptosis-related cysteine peptidase |
| chr12_+_12938541 | 0.10 |
ENST00000356591.4 |
APOLD1 |
apolipoprotein L domain containing 1 |
| chr3_-_183735651 | 0.10 |
ENST00000427120.2 ENST00000392579.2 ENST00000382494.2 ENST00000446941.2 |
ABCC5 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr11_+_64053005 | 0.10 |
ENST00000538032.1 |
GPR137 |
G protein-coupled receptor 137 |
| chr1_+_61869748 | 0.10 |
ENST00000357977.5 |
NFIA |
nuclear factor I/A |
| chr7_-_27169801 | 0.10 |
ENST00000511914.1 |
HOXA4 |
homeobox A4 |
| chr13_-_74708372 | 0.09 |
ENST00000377666.4 |
KLF12 |
Kruppel-like factor 12 |
| chr15_+_40697988 | 0.09 |
ENST00000487418.2 ENST00000479013.2 |
IVD |
isovaleryl-CoA dehydrogenase |
| chr10_+_112836779 | 0.09 |
ENST00000280155.2 |
ADRA2A |
adrenoceptor alpha 2A |
| chr7_+_90339169 | 0.09 |
ENST00000436577.2 |
CDK14 |
cyclin-dependent kinase 14 |
| chr1_+_113933371 | 0.09 |
ENST00000369617.4 |
MAGI3 |
membrane associated guanylate kinase, WW and PDZ domain containing 3 |
| chr10_+_124320156 | 0.08 |
ENST00000338354.3 ENST00000344338.3 ENST00000330163.4 ENST00000368909.3 ENST00000368955.3 ENST00000368956.2 |
DMBT1 |
deleted in malignant brain tumors 1 |
| chr17_-_36891830 | 0.08 |
ENST00000578487.1 |
PCGF2 |
polycomb group ring finger 2 |
| chr1_+_61547405 | 0.08 |
ENST00000371189.4 |
NFIA |
nuclear factor I/A |
| chr10_-_33624002 | 0.08 |
ENST00000432372.2 |
NRP1 |
neuropilin 1 |
| chr14_+_51955831 | 0.08 |
ENST00000356218.4 |
FRMD6 |
FERM domain containing 6 |
| chr7_-_120498357 | 0.08 |
ENST00000415871.1 ENST00000222747.3 ENST00000430985.1 |
TSPAN12 |
tetraspanin 12 |
| chr1_+_210406121 | 0.08 |
ENST00000367012.3 |
SERTAD4 |
SERTA domain containing 4 |
| chr3_-_100712292 | 0.08 |
ENST00000495063.1 ENST00000530539.1 |
ABI3BP |
ABI family, member 3 (NESH) binding protein |
| chr19_-_50529193 | 0.07 |
ENST00000596445.1 ENST00000599538.1 |
VRK3 |
vaccinia related kinase 3 |
| chr10_+_124320195 | 0.07 |
ENST00000359586.6 |
DMBT1 |
deleted in malignant brain tumors 1 |
| chr16_-_30102547 | 0.07 |
ENST00000279386.2 |
TBX6 |
T-box 6 |
| chr11_+_64052944 | 0.07 |
ENST00000535675.1 ENST00000543383.1 |
GPR137 |
G protein-coupled receptor 137 |
| chr10_+_115439699 | 0.07 |
ENST00000369315.1 |
CASP7 |
caspase 7, apoptosis-related cysteine peptidase |
| chr18_+_3451584 | 0.07 |
ENST00000551541.1 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr3_+_159570722 | 0.07 |
ENST00000482804.1 |
SCHIP1 |
schwannomin interacting protein 1 |
| chr5_-_88119580 | 0.07 |
ENST00000539796.1 |
MEF2C |
myocyte enhancer factor 2C |
| chr14_-_53258314 | 0.07 |
ENST00000216410.3 ENST00000557604.1 |
GNPNAT1 |
glucosamine-phosphate N-acetyltransferase 1 |
| chr12_-_4758159 | 0.07 |
ENST00000545990.2 |
AKAP3 |
A kinase (PRKA) anchor protein 3 |
| chr10_+_74927875 | 0.07 |
ENST00000242505.6 |
FAM149B1 |
family with sequence similarity 149, member B1 |
| chr21_+_38792602 | 0.07 |
ENST00000398960.2 ENST00000398956.2 |
DYRK1A |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A |
| chrX_+_106045891 | 0.07 |
ENST00000357242.5 ENST00000310452.2 ENST00000481617.2 ENST00000276175.3 |
TBC1D8B |
TBC1 domain family, member 8B (with GRAM domain) |
| chr9_-_104357277 | 0.07 |
ENST00000374806.1 |
PPP3R2 |
protein phosphatase 3, regulatory subunit B, beta |
| chr16_+_23313591 | 0.07 |
ENST00000343070.2 |
SCNN1B |
sodium channel, non-voltage-gated 1, beta subunit |
| chrX_-_46187069 | 0.07 |
ENST00000446884.1 |
RP1-30G7.2 |
RP1-30G7.2 |
| chr10_-_33623826 | 0.06 |
ENST00000374867.2 |
NRP1 |
neuropilin 1 |
| chr15_+_67418047 | 0.06 |
ENST00000540846.2 |
SMAD3 |
SMAD family member 3 |
| chr9_-_98269481 | 0.06 |
ENST00000418258.1 ENST00000553011.1 ENST00000551845.1 |
PTCH1 |
patched 1 |
| chr19_-_39390440 | 0.06 |
ENST00000249396.7 ENST00000414941.1 ENST00000392081.2 |
SIRT2 |
sirtuin 2 |
| chr10_-_33623310 | 0.06 |
ENST00000395995.1 ENST00000374823.5 ENST00000374821.5 ENST00000374816.3 |
NRP1 |
neuropilin 1 |
| chr16_+_85645007 | 0.06 |
ENST00000405402.2 |
GSE1 |
Gse1 coiled-coil protein |
| chr19_-_39390350 | 0.06 |
ENST00000447739.1 ENST00000358931.5 ENST00000407552.1 |
SIRT2 |
sirtuin 2 |
| chr2_+_204193101 | 0.06 |
ENST00000430418.1 ENST00000424558.1 ENST00000261016.6 |
ABI2 |
abl-interactor 2 |
| chr5_+_42423872 | 0.06 |
ENST00000230882.4 ENST00000357703.3 |
GHR |
growth hormone receptor |
| chr1_+_150980889 | 0.06 |
ENST00000450884.1 ENST00000271620.3 ENST00000271619.8 ENST00000368937.1 ENST00000431193.1 ENST00000368936.1 |
PRUNE |
prune exopolyphosphatase |
| chr6_+_33176265 | 0.06 |
ENST00000374656.4 |
RING1 |
ring finger protein 1 |
| chr1_+_150980989 | 0.06 |
ENST00000368935.1 |
PRUNE |
prune exopolyphosphatase |
| chr4_+_26321284 | 0.05 |
ENST00000506956.1 ENST00000512671.1 ENST00000345843.3 ENST00000342295.1 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
| chr14_-_24701539 | 0.05 |
ENST00000534348.1 ENST00000524927.1 ENST00000250495.5 |
NEDD8-MDP1 NEDD8 |
NEDD8-MDP1 readthrough neural precursor cell expressed, developmentally down-regulated 8 |
| chr5_+_175815732 | 0.05 |
ENST00000274787.2 |
HIGD2A |
HIG1 hypoxia inducible domain family, member 2A |
| chr17_-_16118835 | 0.05 |
ENST00000582357.1 ENST00000436828.1 ENST00000411510.1 ENST00000268712.3 |
NCOR1 |
nuclear receptor corepressor 1 |
| chr10_-_33623564 | 0.05 |
ENST00000374875.1 ENST00000374822.4 |
NRP1 |
neuropilin 1 |
| chr8_-_22089533 | 0.05 |
ENST00000321613.3 |
PHYHIP |
phytanoyl-CoA 2-hydroxylase interacting protein |
| chr10_-_27530997 | 0.05 |
ENST00000375901.1 ENST00000412279.1 ENST00000375905.4 |
ACBD5 |
acyl-CoA binding domain containing 5 |
| chr1_-_180471947 | 0.05 |
ENST00000367595.3 |
ACBD6 |
acyl-CoA binding domain containing 6 |
| chr15_+_75182346 | 0.05 |
ENST00000569931.1 ENST00000352410.4 ENST00000566377.1 ENST00000569233.1 ENST00000567132.1 ENST00000564633.1 ENST00000568907.1 ENST00000563422.1 ENST00000564003.1 ENST00000562800.1 ENST00000563786.1 ENST00000535694.1 ENST00000323744.6 ENST00000568828.1 ENST00000562606.1 ENST00000565576.1 ENST00000567570.1 |
MPI |
mannose phosphate isomerase |
| chr11_-_64512273 | 0.05 |
ENST00000377497.3 ENST00000377487.1 ENST00000430645.1 |
RASGRP2 |
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr6_-_26056695 | 0.05 |
ENST00000343677.2 |
HIST1H1C |
histone cluster 1, H1c |
| chr1_-_43855479 | 0.05 |
ENST00000290663.6 ENST00000372457.4 |
MED8 |
mediator complex subunit 8 |
| chr3_-_19975665 | 0.05 |
ENST00000295824.9 ENST00000389256.4 |
EFHB |
EF-hand domain family, member B |
| chr1_+_61547894 | 0.04 |
ENST00000403491.3 |
NFIA |
nuclear factor I/A |
| chr11_+_64053311 | 0.04 |
ENST00000540370.1 |
GPR137 |
G protein-coupled receptor 137 |
| chr10_-_49459800 | 0.04 |
ENST00000305531.3 |
FRMPD2 |
FERM and PDZ domain containing 2 |
| chr20_+_32250079 | 0.04 |
ENST00000375222.3 |
C20orf144 |
chromosome 20 open reading frame 144 |
| chr1_-_150980828 | 0.04 |
ENST00000361936.5 ENST00000361738.6 |
FAM63A |
family with sequence similarity 63, member A |
| chr13_-_20357110 | 0.04 |
ENST00000427943.1 |
PSPC1 |
paraspeckle component 1 |
| chr19_+_44100632 | 0.04 |
ENST00000533118.1 |
ZNF576 |
zinc finger protein 576 |
| chr2_+_219824357 | 0.04 |
ENST00000302625.4 |
CDK5R2 |
cyclin-dependent kinase 5, regulatory subunit 2 (p39) |
| chr22_-_31885727 | 0.04 |
ENST00000330125.5 ENST00000344710.5 ENST00000397518.1 |
EIF4ENIF1 |
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr10_-_51371321 | 0.04 |
ENST00000602930.1 |
AGAP8 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 8 |
| chr12_-_90049828 | 0.04 |
ENST00000261173.2 ENST00000348959.3 |
ATP2B1 |
ATPase, Ca++ transporting, plasma membrane 1 |
| chr14_+_24701628 | 0.04 |
ENST00000355299.4 ENST00000559836.1 |
GMPR2 |
guanosine monophosphate reductase 2 |
| chr11_+_111789580 | 0.04 |
ENST00000278601.5 |
C11orf52 |
chromosome 11 open reading frame 52 |
| chr13_-_38443860 | 0.04 |
ENST00000426868.2 ENST00000379681.3 ENST00000338947.5 ENST00000355779.2 ENST00000358477.2 ENST00000379673.2 |
TRPC4 |
transient receptor potential cation channel, subfamily C, member 4 |
| chr11_-_32452357 | 0.03 |
ENST00000379079.2 ENST00000530998.1 |
WT1 |
Wilms tumor 1 |
| chr17_-_71410794 | 0.03 |
ENST00000424778.1 |
SDK2 |
sidekick cell adhesion molecule 2 |
| chr9_-_35691017 | 0.03 |
ENST00000378292.3 |
TPM2 |
tropomyosin 2 (beta) |
| chr12_-_49259643 | 0.03 |
ENST00000309739.5 |
RND1 |
Rho family GTPase 1 |
| chr20_+_30225682 | 0.03 |
ENST00000376075.3 |
COX4I2 |
cytochrome c oxidase subunit IV isoform 2 (lung) |
| chr12_-_90049878 | 0.03 |
ENST00000359142.3 |
ATP2B1 |
ATPase, Ca++ transporting, plasma membrane 1 |
| chr17_-_66097610 | 0.03 |
ENST00000584047.1 ENST00000579629.1 |
AC145343.2 |
AC145343.2 |
| chr19_+_37178482 | 0.03 |
ENST00000536254.2 |
ZNF567 |
zinc finger protein 567 |
| chr1_-_146644122 | 0.03 |
ENST00000254101.3 |
PRKAB2 |
protein kinase, AMP-activated, beta 2 non-catalytic subunit |
| chr9_-_130712995 | 0.03 |
ENST00000373084.4 |
FAM102A |
family with sequence similarity 102, member A |
| chr2_+_150187020 | 0.03 |
ENST00000334166.4 |
LYPD6 |
LY6/PLAUR domain containing 6 |
| chr1_+_8021954 | 0.03 |
ENST00000377491.1 ENST00000377488.1 |
PARK7 |
parkinson protein 7 |
| chr6_+_43968306 | 0.03 |
ENST00000442114.2 ENST00000336600.5 ENST00000439969.2 |
C6orf223 |
chromosome 6 open reading frame 223 |
| chr8_-_101348408 | 0.03 |
ENST00000519527.1 ENST00000522369.1 |
RNF19A |
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr2_+_187350973 | 0.03 |
ENST00000544130.1 |
ZC3H15 |
zinc finger CCCH-type containing 15 |
| chr17_+_41322483 | 0.03 |
ENST00000341165.6 ENST00000586650.1 ENST00000422280.1 |
NBR1 |
neighbor of BRCA1 gene 1 |
| chrX_-_153095945 | 0.03 |
ENST00000164640.4 |
PDZD4 |
PDZ domain containing 4 |
| chr5_+_85913721 | 0.03 |
ENST00000247655.3 ENST00000509578.1 ENST00000515763.1 |
COX7C |
cytochrome c oxidase subunit VIIc |
| chr3_-_45957088 | 0.03 |
ENST00000539217.1 |
LZTFL1 |
leucine zipper transcription factor-like 1 |
| chr8_-_67974552 | 0.02 |
ENST00000357849.4 |
COPS5 |
COP9 signalosome subunit 5 |
| chr6_+_126070726 | 0.02 |
ENST00000368364.3 |
HEY2 |
hes-related family bHLH transcription factor with YRPW motif 2 |
| chr8_-_22089845 | 0.02 |
ENST00000454243.2 |
PHYHIP |
phytanoyl-CoA 2-hydroxylase interacting protein |
| chr11_-_64512803 | 0.02 |
ENST00000377489.1 ENST00000354024.3 |
RASGRP2 |
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr2_-_164592497 | 0.02 |
ENST00000333129.3 ENST00000409634.1 |
FIGN |
fidgetin |
| chrX_+_37865804 | 0.02 |
ENST00000297875.2 ENST00000357972.5 |
SYTL5 |
synaptotagmin-like 5 |
| chr4_-_73935409 | 0.02 |
ENST00000507544.2 ENST00000295890.4 |
COX18 |
COX18 cytochrome C oxidase assembly factor |
| chr12_-_31945172 | 0.02 |
ENST00000340398.3 |
H3F3C |
H3 histone, family 3C |
| chr3_-_52029958 | 0.02 |
ENST00000294189.6 |
RPL29 |
ribosomal protein L29 |
| chr11_+_112832133 | 0.02 |
ENST00000524665.1 |
NCAM1 |
neural cell adhesion molecule 1 |
| chr19_-_40724246 | 0.02 |
ENST00000311308.6 |
TTC9B |
tetratricopeptide repeat domain 9B |
| chr8_+_107738343 | 0.02 |
ENST00000521592.1 |
OXR1 |
oxidation resistance 1 |
| chr16_+_72459838 | 0.02 |
ENST00000564508.1 |
AC004158.3 |
AC004158.3 |
| chr1_+_43855560 | 0.02 |
ENST00000562955.1 |
SZT2 |
seizure threshold 2 homolog (mouse) |
| chr1_-_43855444 | 0.02 |
ENST00000372455.4 |
MED8 |
mediator complex subunit 8 |
| chr1_+_110026544 | 0.02 |
ENST00000369870.3 |
ATXN7L2 |
ataxin 7-like 2 |
| chr5_+_137514687 | 0.02 |
ENST00000394894.3 |
KIF20A |
kinesin family member 20A |
| chr9_+_72658490 | 0.02 |
ENST00000377182.4 |
MAMDC2 |
MAM domain containing 2 |
| chr19_+_44100544 | 0.02 |
ENST00000391965.2 ENST00000525771.1 |
ZNF576 |
zinc finger protein 576 |
| chr1_-_2461684 | 0.01 |
ENST00000378453.3 |
HES5 |
hes family bHLH transcription factor 5 |
| chr5_+_137514834 | 0.01 |
ENST00000508792.1 ENST00000504621.1 |
KIF20A |
kinesin family member 20A |
| chr1_+_43855545 | 0.01 |
ENST00000372450.4 ENST00000310739.4 |
SZT2 |
seizure threshold 2 homolog (mouse) |
| chr12_-_7848364 | 0.01 |
ENST00000329913.3 |
GDF3 |
growth differentiation factor 3 |
| chr11_-_67290867 | 0.01 |
ENST00000353903.5 ENST00000294288.4 |
CABP2 |
calcium binding protein 2 |
| chr10_-_99205607 | 0.01 |
ENST00000477692.2 ENST00000485122.2 ENST00000370886.5 ENST00000370885.4 ENST00000370902.3 ENST00000370884.5 |
EXOSC1 |
exosome component 1 |
| chr16_-_70719925 | 0.01 |
ENST00000338779.6 |
MTSS1L |
metastasis suppressor 1-like |
| chr5_-_137514333 | 0.01 |
ENST00000411594.2 ENST00000430331.1 |
BRD8 |
bromodomain containing 8 |
| chr1_-_197744390 | 0.01 |
ENST00000367396.3 |
DENND1B |
DENN/MADD domain containing 1B |
| chr15_-_70390213 | 0.01 |
ENST00000557997.1 ENST00000317509.8 ENST00000442299.2 |
TLE3 |
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
| chr1_+_50571949 | 0.01 |
ENST00000357083.4 |
ELAVL4 |
ELAV like neuron-specific RNA binding protein 4 |
| chr7_+_102553430 | 0.01 |
ENST00000339431.4 ENST00000249377.4 |
LRRC17 |
leucine rich repeat containing 17 |
| chr16_+_30075463 | 0.01 |
ENST00000562168.1 ENST00000569545.1 |
ALDOA |
aldolase A, fructose-bisphosphate |
| chr3_+_107241783 | 0.01 |
ENST00000415149.2 ENST00000402543.1 ENST00000325805.8 ENST00000427402.1 |
BBX |
bobby sox homolog (Drosophila) |
| chr12_-_14133053 | 0.01 |
ENST00000609686.1 |
GRIN2B |
glutamate receptor, ionotropic, N-methyl D-aspartate 2B |
| chr17_-_34257771 | 0.01 |
ENST00000394529.3 ENST00000293273.6 |
RDM1 |
RAD52 motif 1 |
| chr5_-_137514288 | 0.01 |
ENST00000454473.1 ENST00000418329.1 ENST00000455658.2 ENST00000230901.5 ENST00000402931.1 |
BRD8 |
bromodomain containing 8 |
| chr5_+_63461642 | 0.01 |
ENST00000296615.6 ENST00000381081.2 ENST00000389100.4 |
RNF180 |
ring finger protein 180 |
| chrX_-_118925600 | 0.01 |
ENST00000361575.3 |
RPL39 |
ribosomal protein L39 |
| chr6_+_30297306 | 0.01 |
ENST00000420746.1 ENST00000513556.1 |
TRIM39 TRIM39-RPP21 |
tripartite motif containing 39 TRIM39-RPP21 readthrough |
| chr7_+_100187196 | 0.01 |
ENST00000468962.1 ENST00000427939.2 |
FBXO24 |
F-box protein 24 |
| chr17_-_27053216 | 0.01 |
ENST00000292090.3 |
TLCD1 |
TLC domain containing 1 |
| chr16_+_30075595 | 0.01 |
ENST00000563060.2 |
ALDOA |
aldolase A, fructose-bisphosphate |
| chr14_+_22337014 | 0.01 |
ENST00000390436.2 |
TRAV13-1 |
T cell receptor alpha variable 13-1 |
| chr19_-_46088068 | 0.01 |
ENST00000263275.4 ENST00000323060.3 |
OPA3 |
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) |
| chr1_+_185126291 | 0.01 |
ENST00000367500.4 |
SWT1 |
SWT1 RNA endoribonuclease homolog (S. cerevisiae) |
| chr6_+_20403997 | 0.01 |
ENST00000535432.1 |
E2F3 |
E2F transcription factor 3 |
| chrX_+_36065053 | 0.01 |
ENST00000313548.4 |
CHDC2 |
calponin homology domain containing 2 |
| chr14_-_48144157 | 0.01 |
ENST00000399232.2 |
MDGA2 |
MAM domain containing glycosylphosphatidylinositol anchor 2 |
| chr3_+_107241882 | 0.01 |
ENST00000416476.2 |
BBX |
bobby sox homolog (Drosophila) |
| chr16_+_75182376 | 0.01 |
ENST00000570010.1 ENST00000568079.1 ENST00000464850.1 ENST00000332307.4 ENST00000393430.2 |
ZFP1 |
ZFP1 zinc finger protein |
| chr12_+_4918342 | 0.01 |
ENST00000280684.3 ENST00000433855.1 |
KCNA6 |
potassium voltage-gated channel, shaker-related subfamily, member 6 |
| chr19_+_44100727 | 0.01 |
ENST00000528387.1 ENST00000529930.1 ENST00000336564.4 ENST00000607544.1 ENST00000526798.1 |
ZNF576 SRRM5 |
zinc finger protein 576 serine/arginine repetitive matrix 5 |
| chr16_+_21964662 | 0.00 |
ENST00000561553.1 ENST00000565331.1 |
UQCRC2 |
ubiquinol-cytochrome c reductase core protein II |
| chr1_-_235667716 | 0.00 |
ENST00000313984.3 ENST00000366600.3 |
B3GALNT2 |
beta-1,3-N-acetylgalactosaminyltransferase 2 |
| chr19_+_5681153 | 0.00 |
ENST00000579559.1 ENST00000577222.1 |
HSD11B1L RPL36 |
hydroxysteroid (11-beta) dehydrogenase 1-like ribosomal protein L36 |
| chr17_-_34257731 | 0.00 |
ENST00000431884.2 ENST00000425909.3 ENST00000394528.3 ENST00000430160.2 |
RDM1 |
RAD52 motif 1 |
| chr22_+_20861858 | 0.00 |
ENST00000414658.1 ENST00000432052.1 ENST00000425759.2 ENST00000292733.7 ENST00000542773.1 ENST00000263205.7 ENST00000406969.1 ENST00000382974.2 |
MED15 |
mediator complex subunit 15 |
| chr17_+_29421900 | 0.00 |
ENST00000358273.4 ENST00000356175.3 |
NF1 |
neurofibromin 1 |
| chr17_-_39637392 | 0.00 |
ENST00000246639.2 ENST00000393989.1 |
KRT35 |
keratin 35 |
| chr10_-_97200772 | 0.00 |
ENST00000371241.1 ENST00000354106.3 ENST00000371239.1 ENST00000361941.3 ENST00000277982.5 ENST00000371245.3 |
SORBS1 |
sorbin and SH3 domain containing 1 |
| chr1_+_36690011 | 0.00 |
ENST00000354618.5 ENST00000469141.2 ENST00000478853.1 |
THRAP3 |
thyroid hormone receptor associated protein 3 |
| chr12_+_106976678 | 0.00 |
ENST00000392842.1 |
RFX4 |
regulatory factor X, 4 (influences HLA class II expression) |
| chr1_-_8000872 | 0.00 |
ENST00000377507.3 |
TNFRSF9 |
tumor necrosis factor receptor superfamily, member 9 |
| chr2_+_44001172 | 0.00 |
ENST00000260605.8 ENST00000406852.3 ENST00000443170.3 ENST00000398823.2 ENST00000605786.1 |
DYNC2LI1 |
dynein, cytoplasmic 2, light intermediate chain 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.2 | GO:1904835 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.0 | 0.1 | GO:2000777 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.0 | 0.1 | GO:0032811 | negative regulation of epinephrine secretion(GO:0032811) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.0 | 0.2 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.1 | GO:1905068 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.1 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.1 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.0 | 0.1 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.0 | GO:1902958 | neuron intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036482) positive regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902958) regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903383) negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903384) |
| 0.0 | 0.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.0 | GO:0021722 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.0 | 0.2 | GO:1900038 | negative regulation of cellular response to hypoxia(GO:1900038) |
| 0.0 | 0.1 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.0 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.0 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) |
| 0.0 | 0.1 | GO:0035904 | aorta development(GO:0035904) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.1 | GO:0006552 | leucine catabolic process(GO:0006552) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 0.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.1 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.0 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.2 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.2 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.9 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.0 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.0 | 0.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.0 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0005119 | smoothened binding(GO:0005119) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |