Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for RFX3_RFX2
Z-value: 1.23
Transcription factors associated with RFX3_RFX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
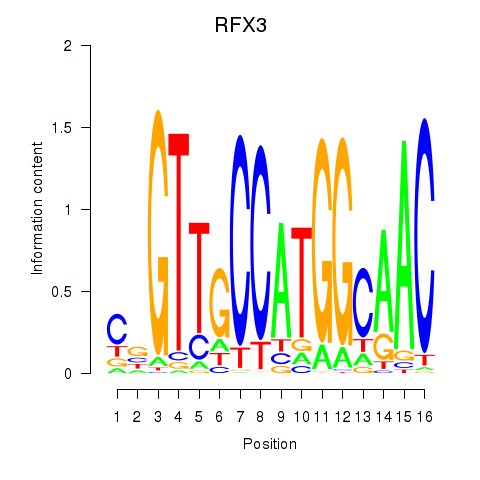
RFX3
|
ENSG00000080298.11 | RFX3 |
|
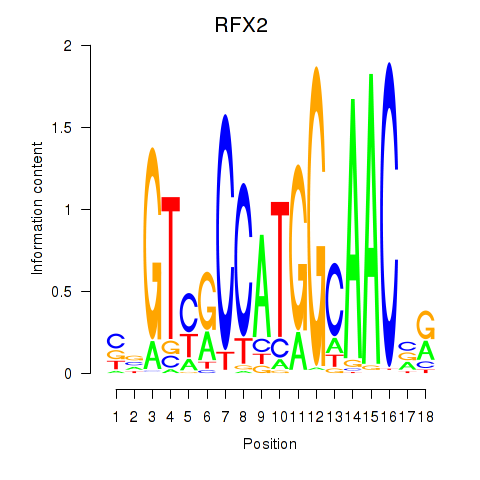
RFX2
|
ENSG00000087903.8 | RFX2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RFX2 | hg19_v2_chr19_-_6110474_6110551 | -0.28 | 5.0e-01 | Click! |
| RFX3 | hg19_v2_chr9_-_3525968_3526016 | 0.21 | 6.2e-01 | Click! |
Activity profile of RFX3_RFX2 motif
Sorted Z-values of RFX3_RFX2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of RFX3_RFX2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_183605200 | 1.63 |
ENST00000304685.4 |
RGL1 |
ral guanine nucleotide dissociation stimulator-like 1 |
| chr5_+_92919043 | 1.58 |
ENST00000327111.3 |
NR2F1 |
nuclear receptor subfamily 2, group F, member 1 |
| chr5_-_179780312 | 1.27 |
ENST00000253778.8 |
GFPT2 |
glutamine-fructose-6-phosphate transaminase 2 |
| chr11_+_133938820 | 1.24 |
ENST00000299106.4 ENST00000529443.2 |
JAM3 |
junctional adhesion molecule 3 |
| chr7_+_100797726 | 1.16 |
ENST00000429457.1 |
AP1S1 |
adaptor-related protein complex 1, sigma 1 subunit |
| chr11_+_133938955 | 1.14 |
ENST00000534549.1 ENST00000441717.3 |
JAM3 |
junctional adhesion molecule 3 |
| chr19_+_35630926 | 1.09 |
ENST00000588081.1 ENST00000589121.1 |
FXYD1 |
FXYD domain containing ion transport regulator 1 |
| chr16_+_8806800 | 1.07 |
ENST00000561870.1 ENST00000396600.2 |
ABAT |
4-aminobutyrate aminotransferase |
| chr12_-_58329819 | 1.06 |
ENST00000551421.1 |
RP11-620J15.3 |
RP11-620J15.3 |
| chr8_+_120428546 | 1.05 |
ENST00000259526.3 |
NOV |
nephroblastoma overexpressed |
| chr14_+_105941118 | 1.04 |
ENST00000550577.1 ENST00000538259.2 |
CRIP2 |
cysteine-rich protein 2 |
| chr10_+_104178946 | 1.01 |
ENST00000432590.1 |
FBXL15 |
F-box and leucine-rich repeat protein 15 |
| chr22_-_43485381 | 0.98 |
ENST00000331018.7 ENST00000266254.7 ENST00000445824.1 |
TTLL1 |
tubulin tyrosine ligase-like family, member 1 |
| chr12_+_4671352 | 0.97 |
ENST00000542744.1 |
DYRK4 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr1_-_232598163 | 0.97 |
ENST00000308942.4 |
SIPA1L2 |
signal-induced proliferation-associated 1 like 2 |
| chr2_+_242811874 | 0.96 |
ENST00000343216.3 |
CXXC11 |
CXXC finger protein 11 |
| chr15_+_43803143 | 0.96 |
ENST00000382031.1 |
MAP1A |
microtubule-associated protein 1A |
| chr19_+_35630022 | 0.95 |
ENST00000589209.1 |
FXYD1 |
FXYD domain containing ion transport regulator 1 |
| chr1_+_245133278 | 0.95 |
ENST00000366522.2 |
EFCAB2 |
EF-hand calcium binding domain 2 |
| chr2_-_175870085 | 0.93 |
ENST00000409156.3 |
CHN1 |
chimerin 1 |
| chr20_+_43160458 | 0.93 |
ENST00000372889.1 ENST00000372887.1 ENST00000372882.3 |
PKIG |
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr20_+_43160409 | 0.91 |
ENST00000372894.3 ENST00000372892.3 ENST00000372891.3 |
PKIG |
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr12_-_63328817 | 0.91 |
ENST00000228705.6 |
PPM1H |
protein phosphatase, Mg2+/Mn2+ dependent, 1H |
| chr2_-_175869936 | 0.89 |
ENST00000409900.3 |
CHN1 |
chimerin 1 |
| chr7_+_150065278 | 0.88 |
ENST00000519397.1 ENST00000479668.1 ENST00000540729.1 |
REPIN1 |
replication initiator 1 |
| chr1_-_109656439 | 0.88 |
ENST00000369949.4 |
C1orf194 |
chromosome 1 open reading frame 194 |
| chr6_+_112408768 | 0.86 |
ENST00000368656.2 ENST00000604268.1 |
FAM229B |
family with sequence similarity 229, member B |
| chr13_-_44361025 | 0.79 |
ENST00000261488.6 |
ENOX1 |
ecto-NOX disulfide-thiol exchanger 1 |
| chr14_+_100259666 | 0.78 |
ENST00000262233.6 ENST00000334192.4 |
EML1 |
echinoderm microtubule associated protein like 1 |
| chr14_+_76044934 | 0.77 |
ENST00000238667.4 |
FLVCR2 |
feline leukemia virus subgroup C cellular receptor family, member 2 |
| chr3_-_58563094 | 0.76 |
ENST00000464064.1 |
FAM107A |
family with sequence similarity 107, member A |
| chrX_+_85969626 | 0.76 |
ENST00000484479.1 |
DACH2 |
dachshund homolog 2 (Drosophila) |
| chr3_-_197676740 | 0.75 |
ENST00000452735.1 ENST00000453254.1 ENST00000455191.1 |
IQCG |
IQ motif containing G |
| chr2_+_74648848 | 0.75 |
ENST00000409791.1 ENST00000426787.1 ENST00000348227.4 |
WDR54 |
WD repeat domain 54 |
| chr16_+_29823427 | 0.74 |
ENST00000358758.7 ENST00000567659.1 ENST00000572820.1 |
PRRT2 |
proline-rich transmembrane protein 2 |
| chr12_-_58329888 | 0.74 |
ENST00000546580.1 |
RP11-620J15.3 |
RP11-620J15.3 |
| chr16_+_29823552 | 0.73 |
ENST00000300797.6 |
PRRT2 |
proline-rich transmembrane protein 2 |
| chr19_+_35630628 | 0.73 |
ENST00000588715.1 ENST00000588607.1 |
FXYD1 |
FXYD domain containing ion transport regulator 1 |
| chr1_+_42928945 | 0.73 |
ENST00000428554.2 |
CCDC30 |
coiled-coil domain containing 30 |
| chr17_-_64187973 | 0.72 |
ENST00000583358.1 ENST00000392769.2 |
CEP112 |
centrosomal protein 112kDa |
| chr1_-_48937838 | 0.72 |
ENST00000371847.3 |
SPATA6 |
spermatogenesis associated 6 |
| chr5_+_148521381 | 0.72 |
ENST00000504238.1 |
ABLIM3 |
actin binding LIM protein family, member 3 |
| chr1_+_38022513 | 0.72 |
ENST00000296218.7 |
DNALI1 |
dynein, axonemal, light intermediate chain 1 |
| chr17_-_64188177 | 0.70 |
ENST00000535342.2 |
CEP112 |
centrosomal protein 112kDa |
| chr22_-_31063782 | 0.70 |
ENST00000404885.1 ENST00000403268.1 ENST00000407308.1 ENST00000342474.4 ENST00000334679.3 |
DUSP18 |
dual specificity phosphatase 18 |
| chr7_+_150065879 | 0.69 |
ENST00000397281.2 ENST00000444957.1 ENST00000466559.1 ENST00000489432.2 ENST00000475514.1 ENST00000482680.1 ENST00000488943.1 ENST00000518514.1 ENST00000478789.1 |
REPIN1 ZNF775 |
replication initiator 1 zinc finger protein 775 |
| chr6_+_30524663 | 0.68 |
ENST00000376560.3 |
PRR3 |
proline rich 3 |
| chr14_+_74486043 | 0.67 |
ENST00000464394.1 ENST00000394009.3 |
CCDC176 |
coiled-coil domain containing 176 |
| chr2_+_220144052 | 0.66 |
ENST00000425450.1 ENST00000392086.4 ENST00000421532.1 |
DNAJB2 |
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr2_-_203735976 | 0.65 |
ENST00000435143.1 |
ICA1L |
islet cell autoantigen 1,69kDa-like |
| chr15_+_55700741 | 0.65 |
ENST00000569691.1 |
C15orf65 |
chromosome 15 open reading frame 65 |
| chr12_-_110486348 | 0.64 |
ENST00000547573.1 ENST00000546651.2 ENST00000551185.2 |
C12orf76 |
chromosome 12 open reading frame 76 |
| chr2_-_130902567 | 0.64 |
ENST00000457413.1 ENST00000392984.3 ENST00000409128.1 ENST00000441670.1 ENST00000409943.3 ENST00000409234.3 ENST00000310463.6 |
CCDC74B |
coiled-coil domain containing 74B |
| chr3_-_196439065 | 0.64 |
ENST00000399942.4 ENST00000409690.3 |
CEP19 |
centrosomal protein 19kDa |
| chrX_+_135251835 | 0.62 |
ENST00000456445.1 |
FHL1 |
four and a half LIM domains 1 |
| chr11_-_6426635 | 0.62 |
ENST00000608645.1 ENST00000608394.1 ENST00000529519.1 |
APBB1 |
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr19_+_10527449 | 0.62 |
ENST00000592685.1 ENST00000380702.2 |
PDE4A |
phosphodiesterase 4A, cAMP-specific |
| chr1_-_48937821 | 0.61 |
ENST00000396199.3 |
SPATA6 |
spermatogenesis associated 6 |
| chr8_+_16884740 | 0.61 |
ENST00000318063.5 |
MICU3 |
mitochondrial calcium uptake family, member 3 |
| chrX_-_71351678 | 0.61 |
ENST00000609883.1 ENST00000545866.1 |
RGAG4 |
retrotransposon gag domain containing 4 |
| chr12_+_51818749 | 0.61 |
ENST00000514353.3 |
SLC4A8 |
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
| chr8_+_13424352 | 0.61 |
ENST00000297324.4 |
C8orf48 |
chromosome 8 open reading frame 48 |
| chr7_+_100797678 | 0.61 |
ENST00000337619.5 |
AP1S1 |
adaptor-related protein complex 1, sigma 1 subunit |
| chr1_+_161068179 | 0.61 |
ENST00000368011.4 ENST00000392192.2 |
KLHDC9 |
kelch domain containing 9 |
| chr6_+_30525051 | 0.61 |
ENST00000376557.3 |
PRR3 |
proline rich 3 |
| chr2_+_132285406 | 0.60 |
ENST00000295171.6 ENST00000409856.3 |
CCDC74A |
coiled-coil domain containing 74A |
| chr12_+_48577366 | 0.60 |
ENST00000316554.3 |
C12orf68 |
chromosome 12 open reading frame 68 |
| chr12_+_51818555 | 0.60 |
ENST00000453097.2 |
SLC4A8 |
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
| chr12_+_51818586 | 0.60 |
ENST00000394856.1 |
SLC4A8 |
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
| chr14_+_24025194 | 0.59 |
ENST00000404535.3 ENST00000288014.6 |
THTPA |
thiamine triphosphatase |
| chr19_+_35630344 | 0.59 |
ENST00000455515.2 |
FXYD1 |
FXYD domain containing ion transport regulator 1 |
| chr14_-_74181106 | 0.56 |
ENST00000316836.3 |
PNMA1 |
paraneoplastic Ma antigen 1 |
| chr10_+_124030819 | 0.55 |
ENST00000260723.4 ENST00000368994.2 |
BTBD16 |
BTB (POZ) domain containing 16 |
| chr19_-_50529193 | 0.54 |
ENST00000596445.1 ENST00000599538.1 |
VRK3 |
vaccinia related kinase 3 |
| chr1_-_48937682 | 0.54 |
ENST00000371843.3 |
SPATA6 |
spermatogenesis associated 6 |
| chr17_-_26662440 | 0.54 |
ENST00000578122.1 |
IFT20 |
intraflagellar transport 20 homolog (Chlamydomonas) |
| chr22_+_27053422 | 0.53 |
ENST00000413665.1 ENST00000421151.1 ENST00000456129.1 ENST00000430080.1 |
MIAT |
myocardial infarction associated transcript (non-protein coding) |
| chr1_+_38022572 | 0.53 |
ENST00000541606.1 |
DNALI1 |
dynein, axonemal, light intermediate chain 1 |
| chr12_-_131323719 | 0.53 |
ENST00000392373.2 |
STX2 |
syntaxin 2 |
| chr3_+_48507210 | 0.53 |
ENST00000433541.1 ENST00000422277.2 ENST00000436480.2 ENST00000444177.1 |
TREX1 |
three prime repair exonuclease 1 |
| chr22_+_27053190 | 0.52 |
ENST00000439738.1 ENST00000422403.1 ENST00000436238.1 ENST00000425476.1 ENST00000455640.1 ENST00000451141.1 ENST00000452429.1 ENST00000423278.1 |
MIAT |
myocardial infarction associated transcript (non-protein coding) |
| chrX_+_103173457 | 0.52 |
ENST00000419165.1 |
TMSB15B |
thymosin beta 15B |
| chr7_-_100965011 | 0.51 |
ENST00000498704.2 ENST00000517481.1 ENST00000437644.2 ENST00000315322.4 |
RABL5 |
RAB, member RAS oncogene family-like 5 |
| chr3_-_196045127 | 0.51 |
ENST00000325318.5 |
TCTEX1D2 |
Tctex1 domain containing 2 |
| chr11_-_66104237 | 0.51 |
ENST00000530056.1 |
RIN1 |
Ras and Rab interactor 1 |
| chr7_+_102073966 | 0.50 |
ENST00000495936.1 ENST00000356387.2 ENST00000478730.2 ENST00000468241.1 ENST00000403646.3 |
ORAI2 |
ORAI calcium release-activated calcium modulator 2 |
| chr17_-_26662464 | 0.50 |
ENST00000579419.1 ENST00000585313.1 ENST00000395418.3 ENST00000578985.1 ENST00000577498.1 ENST00000585089.1 ENST00000357896.3 |
IFT20 |
intraflagellar transport 20 homolog (Chlamydomonas) |
| chr3_+_48507621 | 0.50 |
ENST00000456089.1 |
TREX1 |
three prime repair exonuclease 1 |
| chr17_-_16472483 | 0.50 |
ENST00000395824.1 ENST00000448349.2 ENST00000395825.3 |
ZNF287 |
zinc finger protein 287 |
| chrX_+_135251783 | 0.50 |
ENST00000394153.2 |
FHL1 |
four and a half LIM domains 1 |
| chr12_-_48500085 | 0.49 |
ENST00000549518.1 |
SENP1 |
SUMO1/sentrin specific peptidase 1 |
| chr5_+_154237778 | 0.49 |
ENST00000523698.1 ENST00000517876.1 ENST00000520472.1 |
CNOT8 |
CCR4-NOT transcription complex, subunit 8 |
| chr6_+_52285131 | 0.49 |
ENST00000433625.2 |
EFHC1 |
EF-hand domain (C-terminal) containing 1 |
| chr11_+_111807863 | 0.48 |
ENST00000440460.2 |
DIXDC1 |
DIX domain containing 1 |
| chr5_-_133702761 | 0.47 |
ENST00000521118.1 ENST00000265334.4 ENST00000435211.1 |
CDKL3 |
cyclin-dependent kinase-like 3 |
| chr16_+_3550924 | 0.47 |
ENST00000576634.1 ENST00000574369.1 ENST00000341633.5 ENST00000417763.2 ENST00000571025.1 |
CLUAP1 |
clusterin associated protein 1 |
| chr19_-_46974664 | 0.47 |
ENST00000438932.2 |
PNMAL1 |
paraneoplastic Ma antigen family-like 1 |
| chr19_-_46974741 | 0.47 |
ENST00000313683.10 ENST00000602246.1 |
PNMAL1 |
paraneoplastic Ma antigen family-like 1 |
| chr19_+_3572758 | 0.47 |
ENST00000416526.1 |
HMG20B |
high mobility group 20B |
| chr9_+_35829208 | 0.47 |
ENST00000439587.2 ENST00000377991.4 |
TMEM8B |
transmembrane protein 8B |
| chr17_+_66508537 | 0.46 |
ENST00000392711.1 ENST00000585427.1 ENST00000589228.1 ENST00000536854.2 ENST00000588702.1 ENST00000589309.1 |
PRKAR1A |
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chrX_+_135252050 | 0.45 |
ENST00000449474.1 ENST00000345434.3 |
FHL1 |
four and a half LIM domains 1 |
| chr17_+_66508154 | 0.44 |
ENST00000358598.2 |
PRKAR1A |
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr4_-_186733363 | 0.44 |
ENST00000393523.2 ENST00000393528.3 ENST00000449407.2 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr12_+_56511943 | 0.44 |
ENST00000257940.2 ENST00000552345.1 ENST00000551880.1 ENST00000546903.1 ENST00000551790.1 |
ZC3H10 ESYT1 |
zinc finger CCCH-type containing 10 extended synaptotagmin-like protein 1 |
| chr3_-_107941209 | 0.44 |
ENST00000492106.1 |
IFT57 |
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr9_-_131418944 | 0.44 |
ENST00000419989.1 ENST00000451652.1 ENST00000372715.2 |
WDR34 |
WD repeat domain 34 |
| chr22_+_37447771 | 0.44 |
ENST00000402077.3 ENST00000403888.3 ENST00000456470.1 |
KCTD17 |
potassium channel tetramerization domain containing 17 |
| chr1_-_36916066 | 0.44 |
ENST00000315643.9 |
OSCP1 |
organic solute carrier partner 1 |
| chr12_+_53689309 | 0.43 |
ENST00000351500.3 ENST00000550846.1 ENST00000334478.4 ENST00000549759.1 |
PFDN5 |
prefoldin subunit 5 |
| chr6_-_41703296 | 0.43 |
ENST00000373033.1 |
TFEB |
transcription factor EB |
| chr22_+_39052632 | 0.43 |
ENST00000411557.1 ENST00000396811.2 ENST00000216029.3 ENST00000416285.1 |
CBY1 |
chibby homolog 1 (Drosophila) |
| chr19_-_14048804 | 0.43 |
ENST00000254320.3 ENST00000586075.1 |
PODNL1 |
podocan-like 1 |
| chr19_-_6393216 | 0.42 |
ENST00000595047.1 |
GTF2F1 |
general transcription factor IIF, polypeptide 1, 74kDa |
| chr1_-_118727781 | 0.42 |
ENST00000336338.5 |
SPAG17 |
sperm associated antigen 17 |
| chr6_-_41703952 | 0.42 |
ENST00000358871.2 ENST00000403298.4 |
TFEB |
transcription factor EB |
| chr6_+_52285046 | 0.41 |
ENST00000371068.5 |
EFHC1 |
EF-hand domain (C-terminal) containing 1 |
| chr8_+_38243821 | 0.41 |
ENST00000519476.2 |
LETM2 |
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr12_+_7014064 | 0.41 |
ENST00000443597.2 |
LRRC23 |
leucine rich repeat containing 23 |
| chr16_+_57279248 | 0.41 |
ENST00000562023.1 ENST00000563234.1 |
ARL2BP |
ADP-ribosylation factor-like 2 binding protein |
| chr3_-_16306432 | 0.41 |
ENST00000383775.4 ENST00000488423.1 |
DPH3 |
diphthamide biosynthesis 3 |
| chr3_+_9851632 | 0.41 |
ENST00000426895.4 |
TTLL3 |
tubulin tyrosine ligase-like family, member 3 |
| chr17_+_260097 | 0.41 |
ENST00000360127.6 ENST00000571106.1 ENST00000491373.1 |
C17orf97 |
chromosome 17 open reading frame 97 |
| chr19_+_17413663 | 0.40 |
ENST00000594999.1 |
MRPL34 |
mitochondrial ribosomal protein L34 |
| chr11_+_46958248 | 0.40 |
ENST00000536126.1 ENST00000278460.7 ENST00000378618.2 ENST00000395460.2 ENST00000378615.3 ENST00000543718.1 |
C11orf49 |
chromosome 11 open reading frame 49 |
| chr7_-_44229022 | 0.40 |
ENST00000403799.3 |
GCK |
glucokinase (hexokinase 4) |
| chr2_+_55746746 | 0.40 |
ENST00000406691.3 ENST00000349456.4 ENST00000407816.3 ENST00000403007.3 |
CCDC104 |
coiled-coil domain containing 104 |
| chr17_+_6544078 | 0.40 |
ENST00000250101.5 |
TXNDC17 |
thioredoxin domain containing 17 |
| chr14_+_24025462 | 0.39 |
ENST00000556015.1 ENST00000554970.1 ENST00000554789.1 |
THTPA |
thiamine triphosphatase |
| chr15_-_55700457 | 0.39 |
ENST00000442196.3 ENST00000563171.1 ENST00000425574.3 |
CCPG1 |
cell cycle progression 1 |
| chr7_-_1177874 | 0.39 |
ENST00000397098.3 ENST00000357429.6 ENST00000397100.2 ENST00000491163.1 |
C7orf50 |
chromosome 7 open reading frame 50 |
| chr2_+_39103103 | 0.39 |
ENST00000340556.6 ENST00000410014.1 ENST00000409665.1 ENST00000409077.2 ENST00000409131.2 |
MORN2 |
MORN repeat containing 2 |
| chr12_+_9067123 | 0.38 |
ENST00000543824.1 |
PHC1 |
polyhomeotic homolog 1 (Drosophila) |
| chr5_-_81046904 | 0.38 |
ENST00000515395.1 |
SSBP2 |
single-stranded DNA binding protein 2 |
| chr11_-_66103932 | 0.38 |
ENST00000311320.4 |
RIN1 |
Ras and Rab interactor 1 |
| chr11_+_120894781 | 0.38 |
ENST00000529397.1 ENST00000528512.1 ENST00000422003.2 |
TBCEL |
tubulin folding cofactor E-like |
| chr15_-_65282232 | 0.37 |
ENST00000416889.2 |
SPG21 |
spastic paraplegia 21 (autosomal recessive, Mast syndrome) |
| chr12_+_7013897 | 0.37 |
ENST00000007969.8 ENST00000323702.5 |
LRRC23 |
leucine rich repeat containing 23 |
| chr12_-_110011288 | 0.37 |
ENST00000540016.1 ENST00000266839.5 |
MMAB |
methylmalonic aciduria (cobalamin deficiency) cblB type |
| chr11_+_66360665 | 0.36 |
ENST00000310190.4 |
CCS |
copper chaperone for superoxide dismutase |
| chr21_+_40817749 | 0.36 |
ENST00000380637.3 ENST00000380634.1 ENST00000458295.1 ENST00000440288.2 ENST00000380631.1 |
SH3BGR |
SH3 domain binding glutamic acid-rich protein |
| chr17_-_19266045 | 0.36 |
ENST00000395616.3 |
B9D1 |
B9 protein domain 1 |
| chr2_+_48667983 | 0.36 |
ENST00000449090.2 |
PPP1R21 |
protein phosphatase 1, regulatory subunit 21 |
| chr19_-_14049184 | 0.36 |
ENST00000339560.5 |
PODNL1 |
podocan-like 1 |
| chr15_-_65281775 | 0.36 |
ENST00000433215.2 ENST00000558415.1 ENST00000557795.1 |
SPG21 |
spastic paraplegia 21 (autosomal recessive, Mast syndrome) |
| chr15_-_65282274 | 0.36 |
ENST00000204566.2 |
SPG21 |
spastic paraplegia 21 (autosomal recessive, Mast syndrome) |
| chr3_-_149688896 | 0.36 |
ENST00000239940.7 |
PFN2 |
profilin 2 |
| chr16_+_57279004 | 0.36 |
ENST00000219204.3 |
ARL2BP |
ADP-ribosylation factor-like 2 binding protein |
| chr5_-_159797627 | 0.35 |
ENST00000393975.3 |
C1QTNF2 |
C1q and tumor necrosis factor related protein 2 |
| chr2_+_55746722 | 0.35 |
ENST00000339012.3 |
CCDC104 |
coiled-coil domain containing 104 |
| chr5_-_81046841 | 0.35 |
ENST00000509013.2 ENST00000505980.1 ENST00000509053.1 |
SSBP2 |
single-stranded DNA binding protein 2 |
| chr3_-_50383096 | 0.35 |
ENST00000442887.1 ENST00000360165.3 |
ZMYND10 |
zinc finger, MYND-type containing 10 |
| chrX_-_131623874 | 0.34 |
ENST00000436215.1 |
MBNL3 |
muscleblind-like splicing regulator 3 |
| chr15_-_43802769 | 0.34 |
ENST00000263801.3 |
TP53BP1 |
tumor protein p53 binding protein 1 |
| chr11_-_66103867 | 0.34 |
ENST00000424433.2 |
RIN1 |
Ras and Rab interactor 1 |
| chr1_-_36916011 | 0.34 |
ENST00000356637.5 ENST00000354267.3 ENST00000235532.5 |
OSCP1 |
organic solute carrier partner 1 |
| chr11_-_67271723 | 0.33 |
ENST00000533391.1 ENST00000534749.1 ENST00000532703.1 |
PITPNM1 |
phosphatidylinositol transfer protein, membrane-associated 1 |
| chr17_-_15244894 | 0.33 |
ENST00000338696.2 ENST00000543896.1 ENST00000539245.1 ENST00000539316.1 ENST00000395930.1 |
TEKT3 |
tektin 3 |
| chr13_-_114843416 | 0.33 |
ENST00000389544.4 |
RASA3 |
RAS p21 protein activator 3 |
| chr1_-_114301503 | 0.33 |
ENST00000447664.2 |
PHTF1 |
putative homeodomain transcription factor 1 |
| chr14_-_20774092 | 0.33 |
ENST00000423949.2 ENST00000553828.1 ENST00000258821.3 |
TTC5 |
tetratricopeptide repeat domain 5 |
| chr6_-_27440460 | 0.33 |
ENST00000377419.1 |
ZNF184 |
zinc finger protein 184 |
| chr9_+_100069933 | 0.33 |
ENST00000529487.1 |
CCDC180 |
coiled-coil domain containing 180 |
| chr6_+_35227449 | 0.32 |
ENST00000373953.3 ENST00000440666.2 ENST00000339411.5 |
ZNF76 |
zinc finger protein 76 |
| chr17_-_76899275 | 0.32 |
ENST00000322630.2 ENST00000586713.1 |
DDC8 |
Protein DDC8 homolog |
| chr8_-_101118185 | 0.32 |
ENST00000523437.1 |
RGS22 |
regulator of G-protein signaling 22 |
| chr12_-_54653313 | 0.32 |
ENST00000550411.1 ENST00000439541.2 |
CBX5 |
chromobox homolog 5 |
| chr1_+_3541543 | 0.32 |
ENST00000378344.2 ENST00000344579.5 |
TPRG1L |
tumor protein p63 regulated 1-like |
| chr3_-_49055991 | 0.32 |
ENST00000441576.2 ENST00000420952.2 ENST00000341949.4 ENST00000395462.4 |
DALRD3 |
DALR anticodon binding domain containing 3 |
| chr17_-_72358001 | 0.32 |
ENST00000375366.3 |
BTBD17 |
BTB (POZ) domain containing 17 |
| chr1_+_161129254 | 0.32 |
ENST00000368002.3 ENST00000289865.8 ENST00000479344.1 ENST00000368001.1 |
USP21 |
ubiquitin specific peptidase 21 |
| chr1_+_33546714 | 0.32 |
ENST00000294517.6 ENST00000358680.3 ENST00000373443.3 ENST00000398167.1 |
ADC |
arginine decarboxylase |
| chr3_-_107941230 | 0.31 |
ENST00000264538.3 |
IFT57 |
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr1_-_167905225 | 0.31 |
ENST00000367846.4 |
MPC2 |
mitochondrial pyruvate carrier 2 |
| chr6_-_27440837 | 0.31 |
ENST00000211936.6 |
ZNF184 |
zinc finger protein 184 |
| chr20_-_48532046 | 0.31 |
ENST00000543716.1 |
SPATA2 |
spermatogenesis associated 2 |
| chr17_+_78389247 | 0.31 |
ENST00000520136.2 ENST00000520284.1 ENST00000517795.1 ENST00000523228.1 ENST00000523828.1 ENST00000522200.1 ENST00000521565.1 ENST00000518907.1 ENST00000518644.1 ENST00000518901.1 |
ENDOV |
endonuclease V |
| chr17_-_53046058 | 0.31 |
ENST00000571584.1 ENST00000299335.3 |
COX11 |
cytochrome c oxidase assembly homolog 11 (yeast) |
| chr5_-_81046922 | 0.31 |
ENST00000514493.1 ENST00000320672.4 |
SSBP2 |
single-stranded DNA binding protein 2 |
| chr3_+_9851904 | 0.30 |
ENST00000547186.1 ENST00000397241.1 ENST00000426827.1 |
TTLL3 |
tubulin tyrosine ligase-like family, member 3 |
| chrX_+_48455866 | 0.30 |
ENST00000376729.5 ENST00000218056.5 |
WDR13 |
WD repeat domain 13 |
| chr7_+_138916231 | 0.30 |
ENST00000473989.3 ENST00000288561.8 |
UBN2 |
ubinuclein 2 |
| chr19_+_18263928 | 0.30 |
ENST00000222254.8 |
PIK3R2 |
phosphoinositide-3-kinase, regulatory subunit 2 (beta) |
| chr6_-_43478239 | 0.30 |
ENST00000372441.1 |
LRRC73 |
leucine rich repeat containing 73 |
| chr2_+_237994519 | 0.30 |
ENST00000392008.2 ENST00000409334.1 ENST00000409629.1 |
COPS8 |
COP9 signalosome subunit 8 |
| chr19_-_50528584 | 0.30 |
ENST00000594092.1 ENST00000443401.2 ENST00000594948.1 ENST00000377011.2 ENST00000593919.1 ENST00000601324.1 ENST00000316763.3 ENST00000601341.1 ENST00000600259.1 |
VRK3 |
vaccinia related kinase 3 |
| chr12_-_131323754 | 0.30 |
ENST00000261653.6 |
STX2 |
syntaxin 2 |
| chr19_+_50879705 | 0.30 |
ENST00000598168.1 ENST00000411902.2 ENST00000253727.5 ENST00000597790.1 ENST00000597130.1 ENST00000599105.1 |
NR1H2 |
nuclear receptor subfamily 1, group H, member 2 |
| chr11_-_124981475 | 0.29 |
ENST00000532156.1 ENST00000532407.1 ENST00000279968.4 ENST00000527766.1 ENST00000529583.1 ENST00000524373.1 ENST00000527271.1 ENST00000526175.1 ENST00000529609.1 ENST00000533273.1 ENST00000531909.1 ENST00000529530.1 |
TMEM218 |
transmembrane protein 218 |
| chr22_-_44894178 | 0.29 |
ENST00000341255.3 |
LDOC1L |
leucine zipper, down-regulated in cancer 1-like |
| chr11_+_61447845 | 0.29 |
ENST00000257215.5 |
DAGLA |
diacylglycerol lipase, alpha |
| chr19_-_4535233 | 0.29 |
ENST00000381848.3 ENST00000588887.1 ENST00000586133.1 |
PLIN5 |
perilipin 5 |
| chr19_+_46850251 | 0.29 |
ENST00000012443.4 |
PPP5C |
protein phosphatase 5, catalytic subunit |
| chr19_-_58951496 | 0.28 |
ENST00000254166.3 |
ZNF132 |
zinc finger protein 132 |
| chr16_+_4784458 | 0.28 |
ENST00000590191.1 |
C16orf71 |
chromosome 16 open reading frame 71 |
| chr2_-_230579185 | 0.28 |
ENST00000341772.4 |
DNER |
delta/notch-like EGF repeat containing |
| chr7_+_99699280 | 0.28 |
ENST00000421755.1 |
AP4M1 |
adaptor-related protein complex 4, mu 1 subunit |
| chr12_-_4754339 | 0.28 |
ENST00000228850.1 |
AKAP3 |
A kinase (PRKA) anchor protein 3 |
| chr14_+_102829300 | 0.28 |
ENST00000359520.7 |
TECPR2 |
tectonin beta-propeller repeat containing 2 |
| chr5_+_176873789 | 0.28 |
ENST00000323249.3 ENST00000502922.1 |
PRR7 |
proline rich 7 (synaptic) |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.4 | GO:0010732 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.3 | 1.3 | GO:1904450 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.3 | 1.0 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.2 | 1.0 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.2 | 2.2 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.2 | 0.6 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.2 | 1.9 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.2 | 0.7 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.2 | 0.8 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 2.7 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.8 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 1.5 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 1.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 0.4 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.1 | 0.7 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.1 | 0.3 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.1 | 0.3 | GO:0019364 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.1 | 0.3 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.1 | 1.3 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.1 | 0.3 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.1 | 0.4 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.1 | 0.5 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 2.7 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 1.0 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 0.3 | GO:1902361 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.1 | 0.3 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 0.2 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.1 | 0.3 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.1 | 1.5 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.1 | 0.3 | GO:0098917 | retrograde trans-synaptic signaling(GO:0098917) |
| 0.1 | 0.6 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.3 | GO:0021769 | orbitofrontal cortex development(GO:0021769) |
| 0.1 | 0.3 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.1 | 0.2 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.2 | GO:1901860 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) |
| 0.1 | 0.4 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.1 | 0.4 | GO:0034287 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.1 | 0.7 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 0.1 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.1 | 0.3 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.1 | 0.2 | GO:0061568 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 0.1 | 0.3 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.6 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.7 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.4 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:0014740 | negative regulation of muscle hyperplasia(GO:0014740) |
| 0.0 | 0.1 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.0 | 0.3 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.0 | 0.2 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.0 | 1.4 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.7 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.3 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.7 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.5 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.9 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.0 | 0.4 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.2 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 1.3 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 1.7 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.3 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.5 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.3 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
| 0.0 | 0.7 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.2 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 1.8 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 1.1 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.3 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 1.6 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 0.4 | GO:0046321 | positive regulation of fatty acid oxidation(GO:0046321) |
| 0.0 | 0.8 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.9 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.2 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.0 | 0.3 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.7 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.2 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.0 | 0.2 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.1 | GO:0032425 | positive regulation of mismatch repair(GO:0032425) |
| 0.0 | 0.1 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 | 0.1 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.1 | GO:1902612 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.0 | 0.1 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.5 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 1.1 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.1 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.4 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.4 | GO:0002437 | inflammatory response to antigenic stimulus(GO:0002437) |
| 0.0 | 1.0 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.2 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.4 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.1 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.2 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.8 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:0021678 | third ventricle development(GO:0021678) |
| 0.0 | 0.1 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.5 | GO:0030517 | negative regulation of axon extension(GO:0030517) |
| 0.0 | 1.0 | GO:0018108 | peptidyl-tyrosine phosphorylation(GO:0018108) peptidyl-tyrosine modification(GO:0018212) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 0.8 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.5 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 0.0 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 0.0 | 0.3 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.4 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.9 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.3 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.3 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 | 0.1 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 1.5 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.2 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.3 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.2 | GO:0042424 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.0 | 0.0 | GO:1904637 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.0 | 0.8 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.0 | GO:0070213 | negative regulation of sister chromatid cohesion(GO:0045875) protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.0 | GO:0071362 | cellular response to ether(GO:0071362) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.3 | 1.0 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.3 | 1.3 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.3 | 1.8 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.2 | 0.5 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.2 | 0.5 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.2 | 0.5 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.1 | 2.4 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 3.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.4 | GO:0016234 | inclusion body(GO:0016234) aggresome(GO:0016235) |
| 0.1 | 1.6 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.2 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.1 | 0.5 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 0.5 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 0.6 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.1 | 1.7 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 0.4 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 0.7 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 0.8 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.5 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.1 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.9 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 1.3 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 1.0 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.5 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.3 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.3 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.2 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 1.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.8 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.4 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.4 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.8 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 1.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 1.0 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.4 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.4 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.4 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.9 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.1 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.1 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.2 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.1 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.4 | GO:0035861 | site of double-strand break(GO:0035861) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0003867 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.3 | 0.3 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.3 | 1.6 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.2 | 1.0 | GO:0002046 | opsin binding(GO:0002046) |
| 0.2 | 1.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 1.0 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.1 | 1.9 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 1.0 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 0.7 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.1 | 2.7 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 0.8 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.1 | 0.4 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.1 | 0.3 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.1 | 0.3 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.1 | 1.8 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.8 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.6 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 0.4 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.1 | 0.5 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 1.7 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 0.3 | GO:0052836 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.1 | 0.3 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.1 | 0.3 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 1.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.4 | GO:0019158 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 1.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 3.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.2 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.4 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.8 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.5 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.3 | GO:0050815 | phosphoserine binding(GO:0050815) phosphothreonine binding(GO:0050816) |
| 0.0 | 0.2 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.0 | 0.1 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.0 | 0.3 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.9 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 1.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.6 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.4 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.5 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.4 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.2 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.2 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.4 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.3 | GO:0046935 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.1 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.0 | 0.7 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.2 | GO:1901611 | phosphatidylglycerol binding(GO:1901611) cardiolipin binding(GO:1901612) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.1 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.0 | 0.1 | GO:0034618 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) arginine binding(GO:0034618) |
| 0.0 | 0.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.4 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 1.1 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.9 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.2 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.0 | 0.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.3 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.1 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.3 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 1.0 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.4 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.3 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.6 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.6 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.8 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.4 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.8 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 1.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.7 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.9 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.4 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.0 | 0.7 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 2.2 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 1.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 2.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |