Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
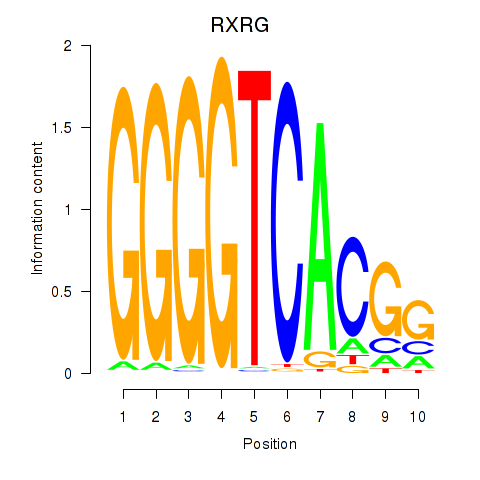
Results for RXRG
Z-value: 0.75
Transcription factors associated with RXRG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RXRG
|
ENSG00000143171.8 | RXRG |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RXRG | hg19_v2_chr1_-_165414414_165414433 | 0.41 | 3.1e-01 | Click! |
Activity profile of RXRG motif
Sorted Z-values of RXRG motif
Network of associatons between targets according to the STRING database.
First level regulatory network of RXRG
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_47074758 | 1.09 |
ENST00000290341.3 |
IGF2BP1 |
insulin-like growth factor 2 mRNA binding protein 1 |
| chr17_+_47075023 | 0.86 |
ENST00000431824.2 |
IGF2BP1 |
insulin-like growth factor 2 mRNA binding protein 1 |
| chr10_-_79398127 | 0.53 |
ENST00000372443.1 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr6_-_46459099 | 0.48 |
ENST00000371374.1 |
RCAN2 |
regulator of calcineurin 2 |
| chr7_-_45960850 | 0.46 |
ENST00000381083.4 ENST00000381086.5 ENST00000275521.6 |
IGFBP3 |
insulin-like growth factor binding protein 3 |
| chrX_-_70474377 | 0.45 |
ENST00000373978.1 ENST00000373981.1 |
ZMYM3 |
zinc finger, MYM-type 3 |
| chr12_-_58131931 | 0.45 |
ENST00000547588.1 |
AGAP2 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chrX_+_30671476 | 0.44 |
ENST00000378946.3 ENST00000378943.3 ENST00000378945.3 ENST00000427190.1 ENST00000378941.3 |
GK |
glycerol kinase |
| chr10_+_76586348 | 0.44 |
ENST00000372724.1 ENST00000287239.4 ENST00000372714.1 |
KAT6B |
K(lysine) acetyltransferase 6B |
| chrX_+_149531524 | 0.41 |
ENST00000370401.2 |
MAMLD1 |
mastermind-like domain containing 1 |
| chr12_+_117176090 | 0.40 |
ENST00000257575.4 ENST00000407967.3 ENST00000392549.2 |
RNFT2 |
ring finger protein, transmembrane 2 |
| chr12_+_117176113 | 0.37 |
ENST00000319176.7 |
RNFT2 |
ring finger protein, transmembrane 2 |
| chr1_-_212873267 | 0.37 |
ENST00000243440.1 |
BATF3 |
basic leucine zipper transcription factor, ATF-like 3 |
| chr10_-_79397479 | 0.35 |
ENST00000404771.3 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr10_-_79397740 | 0.34 |
ENST00000372440.1 ENST00000480683.1 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr5_+_156712372 | 0.34 |
ENST00000541131.1 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
| chr10_-_79397391 | 0.34 |
ENST00000286628.8 ENST00000406533.3 ENST00000354353.5 ENST00000404857.1 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr10_-_79398250 | 0.34 |
ENST00000286627.5 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr6_-_132272504 | 0.32 |
ENST00000367976.3 |
CTGF |
connective tissue growth factor |
| chr3_+_12329358 | 0.30 |
ENST00000309576.6 |
PPARG |
peroxisome proliferator-activated receptor gamma |
| chr2_+_56411131 | 0.29 |
ENST00000407595.2 |
CCDC85A |
coiled-coil domain containing 85A |
| chrX_-_135849484 | 0.29 |
ENST00000370620.1 ENST00000535227.1 |
ARHGEF6 |
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr1_+_23037323 | 0.29 |
ENST00000544305.1 ENST00000374630.3 ENST00000400191.3 ENST00000374632.3 |
EPHB2 |
EPH receptor B2 |
| chrX_-_142722897 | 0.27 |
ENST00000338017.4 |
SLITRK4 |
SLIT and NTRK-like family, member 4 |
| chr17_-_8702667 | 0.27 |
ENST00000329805.4 |
MFSD6L |
major facilitator superfamily domain containing 6-like |
| chr12_+_60083118 | 0.26 |
ENST00000261187.4 ENST00000543448.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr1_+_210406121 | 0.26 |
ENST00000367012.3 |
SERTAD4 |
SERTA domain containing 4 |
| chr22_+_38093005 | 0.26 |
ENST00000406386.3 |
TRIOBP |
TRIO and F-actin binding protein |
| chr10_-_49812997 | 0.25 |
ENST00000417912.2 |
ARHGAP22 |
Rho GTPase activating protein 22 |
| chr9_+_37650945 | 0.25 |
ENST00000377765.3 |
FRMPD1 |
FERM and PDZ domain containing 1 |
| chr9_-_110251836 | 0.25 |
ENST00000374672.4 |
KLF4 |
Kruppel-like factor 4 (gut) |
| chr12_+_51785057 | 0.25 |
ENST00000535225.2 |
SLC4A8 |
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
| chr19_+_35630344 | 0.24 |
ENST00000455515.2 |
FXYD1 |
FXYD domain containing ion transport regulator 1 |
| chr1_-_150693318 | 0.24 |
ENST00000442853.1 ENST00000368995.4 ENST00000368993.2 ENST00000361824.2 ENST00000322343.7 |
HORMAD1 |
HORMA domain containing 1 |
| chr9_-_123342415 | 0.24 |
ENST00000349780.4 ENST00000360190.4 ENST00000360822.3 ENST00000359309.3 |
CDK5RAP2 |
CDK5 regulatory subunit associated protein 2 |
| chrX_-_70473957 | 0.24 |
ENST00000373984.3 ENST00000314425.5 ENST00000373982.1 |
ZMYM3 |
zinc finger, MYM-type 3 |
| chr19_-_47922750 | 0.23 |
ENST00000331559.5 |
MEIS3 |
Meis homeobox 3 |
| chr10_-_81205373 | 0.23 |
ENST00000372336.3 |
ZCCHC24 |
zinc finger, CCHC domain containing 24 |
| chr1_-_159894319 | 0.22 |
ENST00000320307.4 |
TAGLN2 |
transgelin 2 |
| chr17_+_9066252 | 0.22 |
ENST00000436734.1 |
NTN1 |
netrin 1 |
| chr17_-_76921459 | 0.22 |
ENST00000262768.7 |
TIMP2 |
TIMP metallopeptidase inhibitor 2 |
| chr11_+_10326612 | 0.22 |
ENST00000534464.1 ENST00000530439.1 ENST00000524948.1 ENST00000528655.1 ENST00000526492.1 ENST00000525063.1 |
ADM |
adrenomedullin |
| chr19_-_1401486 | 0.22 |
ENST00000252288.2 ENST00000447102.3 |
GAMT |
guanidinoacetate N-methyltransferase |
| chr19_+_10123925 | 0.22 |
ENST00000591589.1 ENST00000171214.1 |
RDH8 |
retinol dehydrogenase 8 (all-trans) |
| chr5_-_37249397 | 0.22 |
ENST00000425232.2 ENST00000274258.7 |
C5orf42 |
chromosome 5 open reading frame 42 |
| chrX_+_55478538 | 0.21 |
ENST00000342972.1 |
MAGEH1 |
melanoma antigen family H, 1 |
| chr19_+_926000 | 0.21 |
ENST00000263620.3 |
ARID3A |
AT rich interactive domain 3A (BRIGHT-like) |
| chr1_-_33168336 | 0.21 |
ENST00000373484.3 |
SYNC |
syncoilin, intermediate filament protein |
| chr16_+_2039946 | 0.21 |
ENST00000248121.2 ENST00000568896.1 |
SYNGR3 |
synaptogyrin 3 |
| chr2_-_202316260 | 0.20 |
ENST00000332624.3 |
TRAK2 |
trafficking protein, kinesin binding 2 |
| chrX_+_153672468 | 0.20 |
ENST00000393600.3 |
FAM50A |
family with sequence similarity 50, member A |
| chr17_+_42264556 | 0.19 |
ENST00000319511.6 ENST00000589785.1 ENST00000592825.1 ENST00000589184.1 |
TMUB2 |
transmembrane and ubiquitin-like domain containing 2 |
| chr5_+_133861790 | 0.19 |
ENST00000395003.1 |
PHF15 |
jade family PHD finger 2 |
| chr8_-_11058847 | 0.19 |
ENST00000297303.4 ENST00000416569.2 |
XKR6 |
XK, Kell blood group complex subunit-related family, member 6 |
| chr2_+_202316392 | 0.19 |
ENST00000194530.3 ENST00000392249.2 |
STRADB |
STE20-related kinase adaptor beta |
| chr17_+_42264395 | 0.19 |
ENST00000587989.1 ENST00000590235.1 |
TMUB2 |
transmembrane and ubiquitin-like domain containing 2 |
| chr5_-_131562935 | 0.18 |
ENST00000379104.2 ENST00000379100.2 ENST00000428369.1 |
P4HA2 |
prolyl 4-hydroxylase, alpha polypeptide II |
| chr19_-_10446449 | 0.18 |
ENST00000592439.1 |
ICAM3 |
intercellular adhesion molecule 3 |
| chr14_+_50999744 | 0.18 |
ENST00000441560.2 |
ATL1 |
atlastin GTPase 1 |
| chr5_-_114515734 | 0.18 |
ENST00000514154.1 ENST00000282369.3 |
TRIM36 |
tripartite motif containing 36 |
| chr9_+_95087766 | 0.18 |
ENST00000375587.3 |
CENPP |
centromere protein P |
| chr12_-_96794143 | 0.18 |
ENST00000543119.2 |
CDK17 |
cyclin-dependent kinase 17 |
| chr20_+_33292068 | 0.18 |
ENST00000374810.3 ENST00000374809.2 ENST00000451665.1 |
TP53INP2 |
tumor protein p53 inducible nuclear protein 2 |
| chr1_-_32210275 | 0.18 |
ENST00000440175.2 |
BAI2 |
brain-specific angiogenesis inhibitor 2 |
| chr19_+_35630628 | 0.18 |
ENST00000588715.1 ENST00000588607.1 |
FXYD1 |
FXYD domain containing ion transport regulator 1 |
| chr7_+_99717230 | 0.17 |
ENST00000262932.3 |
CNPY4 |
canopy FGF signaling regulator 4 |
| chr10_+_115438920 | 0.17 |
ENST00000429617.1 ENST00000369331.4 |
CASP7 |
caspase 7, apoptosis-related cysteine peptidase |
| chr11_-_47447767 | 0.17 |
ENST00000530651.1 ENST00000524447.2 ENST00000531051.2 ENST00000526993.1 ENST00000602866.1 |
PSMC3 |
proteasome (prosome, macropain) 26S subunit, ATPase, 3 |
| chr6_+_126112074 | 0.17 |
ENST00000453302.1 ENST00000417494.1 ENST00000229634.9 |
NCOA7 |
nuclear receptor coactivator 7 |
| chr19_-_47922373 | 0.17 |
ENST00000559524.1 ENST00000557833.1 ENST00000558555.1 ENST00000561293.1 ENST00000441740.2 |
MEIS3 |
Meis homeobox 3 |
| chr20_-_17641097 | 0.17 |
ENST00000246043.4 |
RRBP1 |
ribosome binding protein 1 |
| chr5_+_169532896 | 0.17 |
ENST00000306268.6 ENST00000449804.2 |
FOXI1 |
forkhead box I1 |
| chr17_+_39969183 | 0.17 |
ENST00000321562.4 |
FKBP10 |
FK506 binding protein 10, 65 kDa |
| chr8_+_27632083 | 0.17 |
ENST00000519637.1 |
ESCO2 |
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr8_+_26371763 | 0.17 |
ENST00000521913.1 |
DPYSL2 |
dihydropyrimidinase-like 2 |
| chr11_-_47447970 | 0.16 |
ENST00000298852.3 ENST00000530912.1 |
PSMC3 |
proteasome (prosome, macropain) 26S subunit, ATPase, 3 |
| chr8_-_93115445 | 0.16 |
ENST00000523629.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr8_+_27632047 | 0.16 |
ENST00000397418.2 |
ESCO2 |
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr20_+_43538756 | 0.16 |
ENST00000537323.1 ENST00000217073.2 |
PABPC1L |
poly(A) binding protein, cytoplasmic 1-like |
| chr17_+_42264322 | 0.16 |
ENST00000446571.3 ENST00000357984.3 ENST00000538716.2 |
TMUB2 |
transmembrane and ubiquitin-like domain containing 2 |
| chr13_+_114462193 | 0.16 |
ENST00000375353.3 |
TMEM255B |
transmembrane protein 255B |
| chr10_+_95753714 | 0.16 |
ENST00000260766.3 |
PLCE1 |
phospholipase C, epsilon 1 |
| chr22_-_42466782 | 0.16 |
ENST00000396398.3 ENST00000403363.1 ENST00000402937.1 |
NAGA |
N-acetylgalactosaminidase, alpha- |
| chr15_+_41523417 | 0.16 |
ENST00000560397.1 |
CHP1 |
calcineurin-like EF-hand protein 1 |
| chr7_+_116166331 | 0.16 |
ENST00000393468.1 ENST00000393467.1 |
CAV1 |
caveolin 1, caveolae protein, 22kDa |
| chr17_+_39975455 | 0.16 |
ENST00000455106.1 |
FKBP10 |
FK506 binding protein 10, 65 kDa |
| chrX_+_153686614 | 0.15 |
ENST00000369682.3 |
PLXNA3 |
plexin A3 |
| chr5_+_139505520 | 0.15 |
ENST00000333305.3 |
IGIP |
IgA-inducing protein |
| chr19_-_38714847 | 0.15 |
ENST00000420980.2 ENST00000355526.4 |
DPF1 |
D4, zinc and double PHD fingers family 1 |
| chr6_-_2971494 | 0.15 |
ENST00000380539.1 |
SERPINB6 |
serpin peptidase inhibitor, clade B (ovalbumin), member 6 |
| chr14_-_61190754 | 0.15 |
ENST00000216513.4 |
SIX4 |
SIX homeobox 4 |
| chr5_-_132113559 | 0.15 |
ENST00000448933.1 |
SEPT8 |
septin 8 |
| chr1_-_24194771 | 0.15 |
ENST00000374479.3 |
FUCA1 |
fucosidase, alpha-L- 1, tissue |
| chr1_-_1009683 | 0.15 |
ENST00000453464.2 |
RNF223 |
ring finger protein 223 |
| chr2_-_219433014 | 0.15 |
ENST00000418019.1 ENST00000454775.1 ENST00000338465.5 ENST00000415516.1 ENST00000258399.3 |
USP37 |
ubiquitin specific peptidase 37 |
| chr14_-_89883412 | 0.15 |
ENST00000557258.1 |
FOXN3 |
forkhead box N3 |
| chr5_-_55290773 | 0.15 |
ENST00000502326.3 ENST00000381298.2 |
IL6ST |
interleukin 6 signal transducer (gp130, oncostatin M receptor) |
| chr19_+_32896697 | 0.14 |
ENST00000586987.1 |
DPY19L3 |
dpy-19-like 3 (C. elegans) |
| chr5_+_149569520 | 0.14 |
ENST00000230671.2 ENST00000524041.1 |
SLC6A7 |
solute carrier family 6 (neurotransmitter transporter), member 7 |
| chr17_+_39975544 | 0.14 |
ENST00000544340.1 |
FKBP10 |
FK506 binding protein 10, 65 kDa |
| chr1_+_211433275 | 0.14 |
ENST00000367005.4 |
RCOR3 |
REST corepressor 3 |
| chr20_-_56285595 | 0.14 |
ENST00000395816.3 ENST00000347215.4 |
PMEPA1 |
prostate transmembrane protein, androgen induced 1 |
| chr10_+_54074033 | 0.14 |
ENST00000373970.3 |
DKK1 |
dickkopf WNT signaling pathway inhibitor 1 |
| chr19_+_55795493 | 0.14 |
ENST00000309383.1 |
BRSK1 |
BR serine/threonine kinase 1 |
| chr12_-_133532864 | 0.14 |
ENST00000536932.1 ENST00000360187.4 ENST00000392321.3 |
CHFR ZNF605 |
checkpoint with forkhead and ring finger domains, E3 ubiquitin protein ligase zinc finger protein 605 |
| chr2_+_46926326 | 0.14 |
ENST00000394861.2 |
SOCS5 |
suppressor of cytokine signaling 5 |
| chr22_-_36903069 | 0.14 |
ENST00000216187.6 ENST00000423980.1 |
FOXRED2 |
FAD-dependent oxidoreductase domain containing 2 |
| chr17_-_43045439 | 0.13 |
ENST00000253407.3 |
C1QL1 |
complement component 1, q subcomponent-like 1 |
| chrX_-_70474499 | 0.13 |
ENST00000353904.2 |
ZMYM3 |
zinc finger, MYM-type 3 |
| chr2_-_20101385 | 0.13 |
ENST00000431392.1 |
TTC32 |
tetratricopeptide repeat domain 32 |
| chr21_-_47648665 | 0.13 |
ENST00000450351.1 ENST00000522411.1 ENST00000356396.4 ENST00000397728.3 ENST00000457828.2 |
LSS |
lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) |
| chr13_-_95953589 | 0.13 |
ENST00000538287.1 ENST00000376887.4 ENST00000412704.1 ENST00000536256.1 ENST00000431522.1 |
ABCC4 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 4 |
| chr2_+_27301435 | 0.13 |
ENST00000380320.4 |
EMILIN1 |
elastin microfibril interfacer 1 |
| chr1_+_178062855 | 0.13 |
ENST00000448150.3 |
RASAL2 |
RAS protein activator like 2 |
| chr11_+_72281681 | 0.13 |
ENST00000450804.3 |
RP11-169D4.1 |
RP11-169D4.1 |
| chr8_-_26371608 | 0.13 |
ENST00000522362.2 |
PNMA2 |
paraneoplastic Ma antigen 2 |
| chr5_-_81046922 | 0.13 |
ENST00000514493.1 ENST00000320672.4 |
SSBP2 |
single-stranded DNA binding protein 2 |
| chr14_-_89021077 | 0.13 |
ENST00000556564.1 |
PTPN21 |
protein tyrosine phosphatase, non-receptor type 21 |
| chr2_+_109271481 | 0.13 |
ENST00000542845.1 ENST00000393314.2 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr20_-_57582296 | 0.13 |
ENST00000217131.5 |
CTSZ |
cathepsin Z |
| chr3_-_16555150 | 0.13 |
ENST00000334133.4 |
RFTN1 |
raftlin, lipid raft linker 1 |
| chr17_-_66287257 | 0.13 |
ENST00000327268.4 |
SLC16A6 |
solute carrier family 16, member 6 |
| chr8_+_22423219 | 0.13 |
ENST00000523965.1 ENST00000521554.1 |
SORBS3 |
sorbin and SH3 domain containing 3 |
| chr19_-_39330818 | 0.13 |
ENST00000594769.1 ENST00000602021.1 |
AC104534.3 |
Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial |
| chr5_-_59189545 | 0.12 |
ENST00000340635.6 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
| chr19_-_17375541 | 0.12 |
ENST00000252597.3 |
USHBP1 |
Usher syndrome 1C binding protein 1 |
| chr19_+_7587491 | 0.12 |
ENST00000264079.6 |
MCOLN1 |
mucolipin 1 |
| chr10_+_45495898 | 0.12 |
ENST00000298299.3 |
ZNF22 |
zinc finger protein 22 |
| chr5_-_132113083 | 0.12 |
ENST00000296873.7 |
SEPT8 |
septin 8 |
| chr11_+_71938925 | 0.12 |
ENST00000538751.1 |
INPPL1 |
inositol polyphosphate phosphatase-like 1 |
| chr9_-_140009130 | 0.12 |
ENST00000497375.1 ENST00000371579.2 |
DPP7 |
dipeptidyl-peptidase 7 |
| chr16_+_222846 | 0.12 |
ENST00000251595.6 ENST00000397806.1 |
HBA2 |
hemoglobin, alpha 2 |
| chr11_-_68518910 | 0.12 |
ENST00000544963.1 ENST00000443940.2 |
MTL5 |
metallothionein-like 5, testis-specific (tesmin) |
| chr10_+_124768482 | 0.12 |
ENST00000368869.4 ENST00000358776.4 |
ACADSB |
acyl-CoA dehydrogenase, short/branched chain |
| chr5_-_137667526 | 0.12 |
ENST00000503022.1 |
CDC25C |
cell division cycle 25C |
| chr11_-_116662593 | 0.12 |
ENST00000227665.4 |
APOA5 |
apolipoprotein A-V |
| chr7_-_44229022 | 0.12 |
ENST00000403799.3 |
GCK |
glucokinase (hexokinase 4) |
| chr3_-_169587621 | 0.12 |
ENST00000523069.1 ENST00000316428.5 ENST00000264676.5 |
LRRC31 |
leucine rich repeat containing 31 |
| chr6_-_2971792 | 0.12 |
ENST00000380546.3 |
SERPINB6 |
serpin peptidase inhibitor, clade B (ovalbumin), member 6 |
| chr22_+_18593446 | 0.12 |
ENST00000316027.6 |
TUBA8 |
tubulin, alpha 8 |
| chr9_+_6413317 | 0.12 |
ENST00000276893.5 ENST00000381373.3 |
UHRF2 |
ubiquitin-like with PHD and ring finger domains 2, E3 ubiquitin protein ligase |
| chr5_-_134734901 | 0.12 |
ENST00000312469.4 ENST00000423969.2 |
H2AFY |
H2A histone family, member Y |
| chr2_+_109204743 | 0.12 |
ENST00000332345.6 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr2_+_30670077 | 0.12 |
ENST00000466477.1 ENST00000465200.1 ENST00000379509.3 ENST00000319406.4 ENST00000488144.1 ENST00000465538.1 ENST00000309052.4 ENST00000359433.1 |
LCLAT1 |
lysocardiolipin acyltransferase 1 |
| chr2_-_37899323 | 0.12 |
ENST00000295324.3 ENST00000457889.1 |
CDC42EP3 |
CDC42 effector protein (Rho GTPase binding) 3 |
| chr2_-_230786679 | 0.11 |
ENST00000543084.1 ENST00000343290.5 ENST00000389044.4 ENST00000283943.5 |
TRIP12 |
thyroid hormone receptor interactor 12 |
| chr7_+_116165754 | 0.11 |
ENST00000405348.1 |
CAV1 |
caveolin 1, caveolae protein, 22kDa |
| chr15_-_62352570 | 0.11 |
ENST00000261517.5 ENST00000395896.4 ENST00000395898.3 |
VPS13C |
vacuolar protein sorting 13 homolog C (S. cerevisiae) |
| chr5_-_132112921 | 0.11 |
ENST00000378721.4 ENST00000378701.1 |
SEPT8 |
septin 8 |
| chr20_+_11871433 | 0.11 |
ENST00000399006.2 ENST00000405977.1 |
BTBD3 |
BTB (POZ) domain containing 3 |
| chr11_-_116663127 | 0.11 |
ENST00000433069.1 ENST00000542499.1 |
APOA5 |
apolipoprotein A-V |
| chr6_+_3068606 | 0.11 |
ENST00000259808.4 |
RIPK1 |
receptor (TNFRSF)-interacting serine-threonine kinase 1 |
| chr3_+_170075436 | 0.11 |
ENST00000476188.1 ENST00000259119.4 ENST00000426052.2 |
SKIL |
SKI-like oncogene |
| chr15_-_56035177 | 0.11 |
ENST00000389286.4 ENST00000561292.1 |
PRTG |
protogenin |
| chr17_+_4853442 | 0.11 |
ENST00000522301.1 |
ENO3 |
enolase 3 (beta, muscle) |
| chr12_-_54813229 | 0.11 |
ENST00000293379.4 |
ITGA5 |
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr1_+_245133656 | 0.11 |
ENST00000366521.3 |
EFCAB2 |
EF-hand calcium binding domain 2 |
| chr14_+_60716159 | 0.11 |
ENST00000325658.3 |
PPM1A |
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
| chr15_-_101142362 | 0.11 |
ENST00000559577.1 ENST00000561308.1 ENST00000560133.1 ENST00000560941.1 ENST00000559736.1 ENST00000560272.1 |
LINS |
lines homolog (Drosophila) |
| chr17_+_42923686 | 0.11 |
ENST00000591513.1 |
HIGD1B |
HIG1 hypoxia inducible domain family, member 1B |
| chr15_+_63414760 | 0.11 |
ENST00000557972.1 |
LACTB |
lactamase, beta |
| chr1_-_155881156 | 0.10 |
ENST00000539040.1 ENST00000368323.3 |
RIT1 |
Ras-like without CAAX 1 |
| chr12_-_96794330 | 0.10 |
ENST00000261211.3 |
CDK17 |
cyclin-dependent kinase 17 |
| chr11_+_450255 | 0.10 |
ENST00000308020.5 |
PTDSS2 |
phosphatidylserine synthase 2 |
| chr17_-_6947225 | 0.10 |
ENST00000574600.1 ENST00000308009.1 ENST00000447225.1 |
SLC16A11 |
solute carrier family 16, member 11 |
| chr11_+_827553 | 0.10 |
ENST00000528542.2 ENST00000450448.1 |
EFCAB4A |
EF-hand calcium binding domain 4A |
| chr17_-_1531635 | 0.10 |
ENST00000571650.1 |
SLC43A2 |
solute carrier family 43 (amino acid system L transporter), member 2 |
| chr4_-_85887503 | 0.10 |
ENST00000509172.1 ENST00000322366.6 ENST00000295888.4 ENST00000502713.1 |
WDFY3 |
WD repeat and FYVE domain containing 3 |
| chr1_+_145516560 | 0.10 |
ENST00000537888.1 |
PEX11B |
peroxisomal biogenesis factor 11 beta |
| chr5_-_131563501 | 0.10 |
ENST00000401867.1 ENST00000379086.1 ENST00000418055.1 ENST00000453286.1 ENST00000166534.4 |
P4HA2 |
prolyl 4-hydroxylase, alpha polypeptide II |
| chr16_+_29985503 | 0.10 |
ENST00000543033.1 ENST00000279394.3 |
TAOK2 |
TAO kinase 2 |
| chr1_+_155146318 | 0.10 |
ENST00000368385.4 ENST00000545012.1 ENST00000392451.2 ENST00000368383.3 ENST00000368382.1 ENST00000334634.4 |
TRIM46 |
tripartite motif containing 46 |
| chr6_+_24495067 | 0.10 |
ENST00000357578.3 ENST00000546278.1 ENST00000491546.1 |
ALDH5A1 |
aldehyde dehydrogenase 5 family, member A1 |
| chr16_-_31161380 | 0.10 |
ENST00000569305.1 ENST00000418068.2 ENST00000268281.4 |
PRSS36 |
protease, serine, 36 |
| chrX_-_30327495 | 0.10 |
ENST00000453287.1 |
NR0B1 |
nuclear receptor subfamily 0, group B, member 1 |
| chr17_-_56065484 | 0.10 |
ENST00000581208.1 |
VEZF1 |
vascular endothelial zinc finger 1 |
| chr1_+_1115056 | 0.10 |
ENST00000379288.3 |
TTLL10 |
tubulin tyrosine ligase-like family, member 10 |
| chr14_-_77737543 | 0.10 |
ENST00000298352.4 |
NGB |
neuroglobin |
| chr19_+_50919056 | 0.10 |
ENST00000599632.1 |
CTD-2545M3.6 |
CTD-2545M3.6 |
| chr17_-_80291818 | 0.10 |
ENST00000269389.3 ENST00000581691.1 |
SECTM1 |
secreted and transmembrane 1 |
| chr12_-_75603236 | 0.10 |
ENST00000540018.1 |
KCNC2 |
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr5_+_170288856 | 0.10 |
ENST00000523189.1 |
RANBP17 |
RAN binding protein 17 |
| chr10_+_81892347 | 0.10 |
ENST00000372267.2 |
PLAC9 |
placenta-specific 9 |
| chr1_+_26606608 | 0.10 |
ENST00000319041.6 |
SH3BGRL3 |
SH3 domain binding glutamic acid-rich protein like 3 |
| chr5_-_137667459 | 0.10 |
ENST00000415130.2 ENST00000356505.3 ENST00000357274.3 ENST00000348983.3 ENST00000323760.6 |
CDC25C |
cell division cycle 25C |
| chr19_-_50266580 | 0.10 |
ENST00000246801.3 |
TSKS |
testis-specific serine kinase substrate |
| chr1_+_160051319 | 0.10 |
ENST00000368088.3 |
KCNJ9 |
potassium inwardly-rectifying channel, subfamily J, member 9 |
| chr5_-_132113036 | 0.10 |
ENST00000378706.1 |
SEPT8 |
septin 8 |
| chr11_-_615570 | 0.10 |
ENST00000525445.1 ENST00000348655.6 ENST00000397566.1 |
IRF7 |
interferon regulatory factor 7 |
| chr3_-_46904918 | 0.09 |
ENST00000395869.1 |
MYL3 |
myosin, light chain 3, alkali; ventricular, skeletal, slow |
| chr3_-_46904946 | 0.09 |
ENST00000292327.4 |
MYL3 |
myosin, light chain 3, alkali; ventricular, skeletal, slow |
| chr11_-_65430251 | 0.09 |
ENST00000534283.1 ENST00000527749.1 ENST00000533187.1 ENST00000525693.1 ENST00000534558.1 ENST00000532879.1 ENST00000532999.1 |
RELA |
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr5_+_139781393 | 0.09 |
ENST00000360839.2 ENST00000297183.6 ENST00000421134.1 ENST00000394723.3 ENST00000511151.1 |
ANKHD1 |
ankyrin repeat and KH domain containing 1 |
| chr17_+_7905912 | 0.09 |
ENST00000254854.4 |
GUCY2D |
guanylate cyclase 2D, membrane (retina-specific) |
| chr1_+_145516252 | 0.09 |
ENST00000369306.3 |
PEX11B |
peroxisomal biogenesis factor 11 beta |
| chr20_+_62694834 | 0.09 |
ENST00000415602.1 |
TCEA2 |
transcription elongation factor A (SII), 2 |
| chr17_-_40575535 | 0.09 |
ENST00000357037.5 |
PTRF |
polymerase I and transcript release factor |
| chr2_+_95963052 | 0.09 |
ENST00000295225.5 |
KCNIP3 |
Kv channel interacting protein 3, calsenilin |
| chr20_+_30946106 | 0.09 |
ENST00000375687.4 ENST00000542461.1 |
ASXL1 |
additional sex combs like 1 (Drosophila) |
| chr10_+_70320413 | 0.09 |
ENST00000373644.4 |
TET1 |
tet methylcytosine dioxygenase 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.2 | 1.9 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.2 | 0.5 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.1 | 0.4 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.1 | 0.3 | GO:0034059 | response to anoxia(GO:0034059) |
| 0.1 | 0.3 | GO:1903609 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.1 | 0.3 | GO:0014740 | negative regulation of muscle hyperplasia(GO:0014740) |
| 0.1 | 0.2 | GO:0051598 | meiotic DNA double-strand break formation(GO:0042138) meiotic recombination checkpoint(GO:0051598) |
| 0.1 | 0.3 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.1 | 0.4 | GO:0010734 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.1 | 0.2 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.1 | 0.2 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.1 | 0.4 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.3 | GO:0030047 | actin modification(GO:0030047) |
| 0.1 | 0.2 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.2 | GO:0010902 | positive regulation of very-low-density lipoprotein particle remodeling(GO:0010902) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.2 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.0 | 0.1 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 0.0 | 0.3 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.0 | 0.1 | GO:0032242 | regulation of nucleoside transport(GO:0032242) positive regulation of necroptotic process(GO:0060545) |
| 0.0 | 0.2 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.3 | GO:1904783 | positive regulation of NMDA glutamate receptor activity(GO:1904783) |
| 0.0 | 0.1 | GO:0014040 | positive regulation of Schwann cell differentiation(GO:0014040) |
| 0.0 | 0.3 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.3 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.2 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.1 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.0 | 0.1 | GO:1901295 | positive regulation of ephrin receptor signaling pathway(GO:1901189) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.1 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.1 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.1 | GO:0098838 | reduced folate transmembrane transport(GO:0098838) |
| 0.0 | 0.1 | GO:1903490 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.0 | 0.3 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 0.2 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.2 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.0 | 0.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.0 | 0.1 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.0 | 0.1 | GO:1904048 | regulation of spontaneous neurotransmitter secretion(GO:1904048) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.3 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:0009450 | acetate metabolic process(GO:0006083) gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.0 | 0.2 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.2 | GO:0009730 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.2 | GO:0070885 | positive regulation of sodium:proton antiporter activity(GO:0032417) negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.0 | 0.1 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.0 | 0.4 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.2 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.1 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.0 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.2 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.1 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.1 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.1 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.1 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.1 | GO:0018317 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.1 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.1 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.0 | 0.1 | GO:0097676 | cell migration involved in vasculogenesis(GO:0035441) histone H3-K36 dimethylation(GO:0097676) |
| 0.0 | 0.1 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.0 | 0.1 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.1 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.1 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.0 | GO:0070837 | L-ascorbic acid transport(GO:0015882) dehydroascorbic acid transport(GO:0070837) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.0 | 0.1 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.1 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.1 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.0 | GO:0060661 | submandibular salivary gland formation(GO:0060661) hair follicle cell proliferation(GO:0071335) regulation of hair follicle cell proliferation(GO:0071336) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.0 | 0.0 | GO:2000777 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.0 | 0.5 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.0 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.2 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.1 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.1 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.1 | GO:2000756 | regulation of histone acetylation(GO:0035065) regulation of peptidyl-lysine acetylation(GO:2000756) |
| 0.0 | 0.0 | GO:0060974 | neural crest cell migration involved in heart formation(GO:0003147) cell migration involved in heart formation(GO:0060974) anterior neural tube closure(GO:0061713) |
| 0.0 | 0.1 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.3 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.2 | GO:0021637 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.3 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.1 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.0 | 0.0 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 | 0.0 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.0 | 0.0 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.0 | 0.1 | GO:1904526 | regulation of microtubule binding(GO:1904526) |
| 0.0 | 0.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.1 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.1 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.0 | GO:0050894 | determination of affect(GO:0050894) |
| 0.0 | 0.1 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.0 | 0.0 | GO:0098976 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.1 | GO:0061734 | parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) |
| 0.0 | 0.0 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0071159 | NF-kappaB complex(GO:0071159) |
| 0.0 | 0.4 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.4 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.3 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.4 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.1 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.1 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 2.2 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.3 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.1 | GO:0032279 | axonemal microtubule(GO:0005879) asymmetric synapse(GO:0032279) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.9 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 0.4 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.1 | 0.3 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.1 | 0.5 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.5 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.3 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 0.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 2.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.2 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.1 | 0.3 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.0 | 0.1 | GO:0019981 | interleukin-6 receptor activity(GO:0004915) leukemia inhibitory factor receptor activity(GO:0004923) interleukin-6 binding(GO:0019981) |
| 0.0 | 0.4 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.0 | 0.2 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.0 | 0.1 | GO:0035034 | histone acetyltransferase regulator activity(GO:0035034) |
| 0.0 | 0.1 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.0 | 0.3 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.1 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 0.0 | 0.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.3 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.0 | 0.2 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.3 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.1 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.0 | 0.3 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.1 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.0 | 0.1 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.0 | 0.1 | GO:0051500 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.2 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.1 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.5 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.0 | 0.0 | GO:0033300 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) dehydroascorbic acid transporter activity(GO:0033300) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.0 | 0.1 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.0 | GO:0036361 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.0 | 0.0 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.0 | 0.0 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.0 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.1 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.0 | 0.1 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.2 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.0 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.0 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.0 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.1 | GO:0000182 | rDNA binding(GO:0000182) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 1.9 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.0 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 1.8 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |