Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for SIX1_SIX3_SIX2
Z-value: 0.96
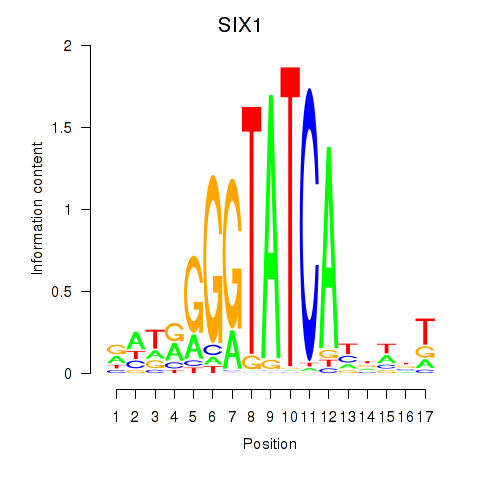
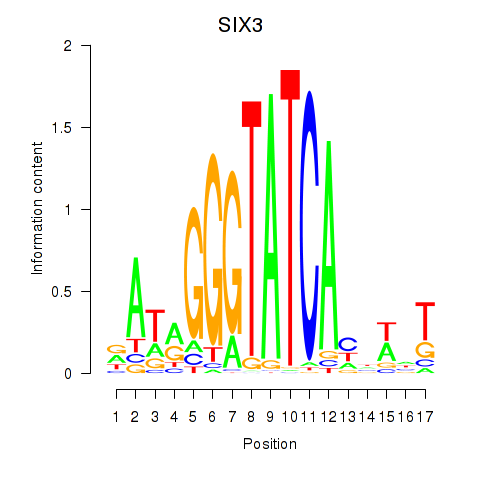
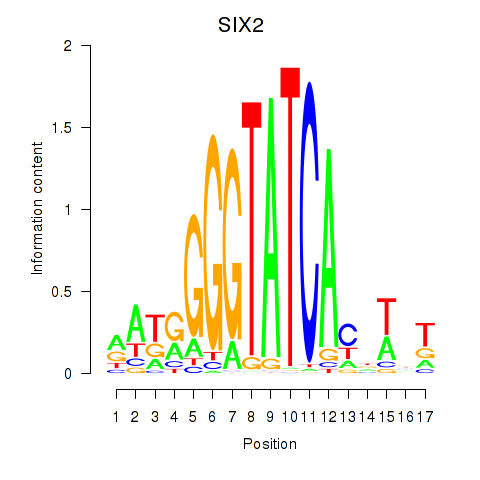
Transcription factors associated with SIX1_SIX3_SIX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SIX1
|
ENSG00000126778.7 | SIX1 |
|
SIX3
|
ENSG00000138083.3 | SIX3 |
|
SIX2
|
ENSG00000170577.7 | SIX2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SIX1 | hg19_v2_chr14_-_61116168_61116180 | -0.71 | 4.7e-02 | Click! |
| SIX3 | hg19_v2_chr2_+_45168875_45168916 | -0.68 | 6.2e-02 | Click! |
| SIX2 | hg19_v2_chr2_-_45236540_45236577 | -0.30 | 4.8e-01 | Click! |
Activity profile of SIX1_SIX3_SIX2 motif
Sorted Z-values of SIX1_SIX3_SIX2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SIX1_SIX3_SIX2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_136847099 | 1.67 |
ENST00000438100.2 |
MAP7 |
microtubule-associated protein 7 |
| chr6_-_136847610 | 1.34 |
ENST00000454590.1 ENST00000432797.2 |
MAP7 |
microtubule-associated protein 7 |
| chr11_-_118134997 | 1.17 |
ENST00000278937.2 |
MPZL2 |
myelin protein zero-like 2 |
| chr11_-_118135160 | 1.07 |
ENST00000438295.2 |
MPZL2 |
myelin protein zero-like 2 |
| chr1_+_15256230 | 0.84 |
ENST00000376028.4 ENST00000400798.2 |
KAZN |
kazrin, periplakin interacting protein |
| chr12_-_58220078 | 0.68 |
ENST00000549039.1 |
CTDSP2 |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr12_-_52845910 | 0.62 |
ENST00000252252.3 |
KRT6B |
keratin 6B |
| chr20_+_44637526 | 0.56 |
ENST00000372330.3 |
MMP9 |
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
| chr6_+_28048753 | 0.50 |
ENST00000377325.1 |
ZNF165 |
zinc finger protein 165 |
| chr10_-_134331695 | 0.49 |
ENST00000455414.1 |
RP11-432J24.5 |
RP11-432J24.5 |
| chr11_-_112034831 | 0.44 |
ENST00000280357.7 |
IL18 |
interleukin 18 (interferon-gamma-inducing factor) |
| chr15_+_40650408 | 0.39 |
ENST00000267889.3 |
DISP2 |
dispatched homolog 2 (Drosophila) |
| chr10_+_48255253 | 0.38 |
ENST00000357718.4 ENST00000344416.5 ENST00000456111.2 ENST00000374258.3 |
ANXA8 AL591684.1 |
annexin A8 Protein LOC100996760 |
| chr2_-_113594279 | 0.36 |
ENST00000416750.1 ENST00000418817.1 ENST00000263341.2 |
IL1B |
interleukin 1, beta |
| chr3_-_111314230 | 0.36 |
ENST00000317012.4 |
ZBED2 |
zinc finger, BED-type containing 2 |
| chr1_+_160709076 | 0.34 |
ENST00000359331.4 ENST00000495334.1 |
SLAMF7 |
SLAM family member 7 |
| chr10_+_47746929 | 0.34 |
ENST00000340243.6 ENST00000374277.5 ENST00000449464.2 ENST00000538825.1 ENST00000335083.5 |
ANXA8L2 AL603965.1 |
annexin A8-like 2 Protein LOC100996760 |
| chr12_+_20848377 | 0.32 |
ENST00000540354.1 ENST00000266509.2 ENST00000381552.1 |
SLCO1C1 |
solute carrier organic anion transporter family, member 1C1 |
| chr1_+_158149737 | 0.31 |
ENST00000368171.3 |
CD1D |
CD1d molecule |
| chr4_-_84255935 | 0.29 |
ENST00000513463.1 |
HPSE |
heparanase |
| chr3_+_111718036 | 0.29 |
ENST00000455401.2 |
TAGLN3 |
transgelin 3 |
| chr22_+_32455111 | 0.28 |
ENST00000543737.1 |
SLC5A1 |
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr1_+_160709055 | 0.28 |
ENST00000368043.3 ENST00000368042.3 ENST00000458602.2 ENST00000458104.2 |
SLAMF7 |
SLAM family member 7 |
| chr3_+_70048881 | 0.27 |
ENST00000483525.1 |
RP11-460N16.1 |
RP11-460N16.1 |
| chr18_+_55816546 | 0.27 |
ENST00000435432.2 ENST00000357895.5 ENST00000586263.1 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr11_+_34654011 | 0.27 |
ENST00000531794.1 |
EHF |
ets homologous factor |
| chr3_+_111718173 | 0.25 |
ENST00000494932.1 |
TAGLN3 |
transgelin 3 |
| chr1_+_196946664 | 0.25 |
ENST00000367414.5 |
CFHR5 |
complement factor H-related 5 |
| chr6_-_138428613 | 0.25 |
ENST00000421351.3 |
PERP |
PERP, TP53 apoptosis effector |
| chr16_+_57673207 | 0.24 |
ENST00000564783.1 ENST00000564729.1 ENST00000565976.1 ENST00000566508.1 ENST00000544297.1 |
GPR56 |
G protein-coupled receptor 56 |
| chr11_+_117947782 | 0.24 |
ENST00000522307.1 ENST00000523251.1 ENST00000437212.3 ENST00000522824.1 ENST00000522151.1 |
TMPRSS4 |
transmembrane protease, serine 4 |
| chr3_+_111717600 | 0.24 |
ENST00000273368.4 |
TAGLN3 |
transgelin 3 |
| chr10_-_47173994 | 0.23 |
ENST00000414655.2 ENST00000545298.1 ENST00000359178.4 ENST00000358140.4 ENST00000503031.1 |
ANXA8L1 LINC00842 |
annexin A8-like 1 long intergenic non-protein coding RNA 842 |
| chr1_+_150245177 | 0.23 |
ENST00000369098.3 |
C1orf54 |
chromosome 1 open reading frame 54 |
| chr3_-_11685345 | 0.23 |
ENST00000430365.2 |
VGLL4 |
vestigial like 4 (Drosophila) |
| chr11_+_117947724 | 0.23 |
ENST00000534111.1 |
TMPRSS4 |
transmembrane protease, serine 4 |
| chr1_-_85462623 | 0.23 |
ENST00000370608.3 |
MCOLN2 |
mucolipin 2 |
| chr4_+_89299994 | 0.22 |
ENST00000264346.7 |
HERC6 |
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr6_-_11779174 | 0.22 |
ENST00000379413.2 |
ADTRP |
androgen-dependent TFPI-regulating protein |
| chr1_+_196946680 | 0.22 |
ENST00000256785.4 |
CFHR5 |
complement factor H-related 5 |
| chr2_-_89247338 | 0.22 |
ENST00000496168.1 |
IGKV1-5 |
immunoglobulin kappa variable 1-5 |
| chr6_-_11779014 | 0.22 |
ENST00000229583.5 |
ADTRP |
androgen-dependent TFPI-regulating protein |
| chr1_-_240906911 | 0.22 |
ENST00000431139.2 |
RP11-80B9.4 |
RP11-80B9.4 |
| chr10_+_5238793 | 0.22 |
ENST00000263126.1 |
AKR1C4 |
aldo-keto reductase family 1, member C4 |
| chr8_+_18248755 | 0.21 |
ENST00000286479.3 |
NAT2 |
N-acetyltransferase 2 (arylamine N-acetyltransferase) |
| chr8_+_18248786 | 0.21 |
ENST00000520116.1 |
NAT2 |
N-acetyltransferase 2 (arylamine N-acetyltransferase) |
| chr14_+_22739823 | 0.21 |
ENST00000390464.2 |
TRAV38-1 |
T cell receptor alpha variable 38-1 |
| chr11_-_34533257 | 0.21 |
ENST00000312319.2 |
ELF5 |
E74-like factor 5 (ets domain transcription factor) |
| chr16_-_28550348 | 0.21 |
ENST00000324873.6 |
NUPR1 |
nuclear protein, transcriptional regulator, 1 |
| chrX_+_56259316 | 0.20 |
ENST00000468660.1 |
KLF8 |
Kruppel-like factor 8 |
| chr3_-_10452359 | 0.20 |
ENST00000452124.1 |
ATP2B2 |
ATPase, Ca++ transporting, plasma membrane 2 |
| chr1_+_26737253 | 0.20 |
ENST00000326279.6 |
LIN28A |
lin-28 homolog A (C. elegans) |
| chr11_-_2924720 | 0.20 |
ENST00000455942.2 |
SLC22A18AS |
solute carrier family 22 (organic cation transporter), member 18 antisense |
| chr9_-_117568365 | 0.20 |
ENST00000374045.4 |
TNFSF15 |
tumor necrosis factor (ligand) superfamily, member 15 |
| chr7_-_141541221 | 0.19 |
ENST00000350549.3 ENST00000438520.1 |
PRSS37 |
protease, serine, 37 |
| chr11_+_100862811 | 0.19 |
ENST00000303130.2 |
TMEM133 |
transmembrane protein 133 |
| chr7_-_121944491 | 0.19 |
ENST00000331178.4 ENST00000427185.2 ENST00000442488.2 |
FEZF1 |
FEZ family zinc finger 1 |
| chr6_+_33043703 | 0.18 |
ENST00000418931.2 ENST00000535465.1 |
HLA-DPB1 |
major histocompatibility complex, class II, DP beta 1 |
| chr16_-_28550320 | 0.18 |
ENST00000395641.2 |
NUPR1 |
nuclear protein, transcriptional regulator, 1 |
| chr1_+_150245099 | 0.18 |
ENST00000369099.3 |
C1orf54 |
chromosome 1 open reading frame 54 |
| chr1_+_161677034 | 0.17 |
ENST00000349527.4 ENST00000309691.6 ENST00000294796.4 ENST00000367953.3 ENST00000367950.1 |
FCRLA |
Fc receptor-like A |
| chr19_-_56249740 | 0.17 |
ENST00000590200.1 ENST00000332836.2 |
NLRP9 |
NLR family, pyrin domain containing 9 |
| chr9_+_12693336 | 0.16 |
ENST00000381137.2 ENST00000388918.5 |
TYRP1 |
tyrosinase-related protein 1 |
| chr1_+_161676983 | 0.16 |
ENST00000367957.2 |
FCRLA |
Fc receptor-like A |
| chr8_-_48651648 | 0.16 |
ENST00000408965.3 |
CEBPD |
CCAAT/enhancer binding protein (C/EBP), delta |
| chr1_+_160709029 | 0.16 |
ENST00000444090.2 ENST00000441662.2 |
SLAMF7 |
SLAM family member 7 |
| chr3_+_145782358 | 0.16 |
ENST00000422482.1 |
AC107021.1 |
HCG1786590; PRO2533; Uncharacterized protein |
| chr11_-_44972390 | 0.15 |
ENST00000395648.3 ENST00000531928.2 |
TP53I11 |
tumor protein p53 inducible protein 11 |
| chr11_-_44972476 | 0.15 |
ENST00000527685.1 ENST00000308212.5 |
TP53I11 |
tumor protein p53 inducible protein 11 |
| chr6_+_116782527 | 0.15 |
ENST00000368606.3 ENST00000368605.1 |
FAM26F |
family with sequence similarity 26, member F |
| chr4_-_153332886 | 0.15 |
ENST00000603841.1 |
FBXW7 |
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr1_+_26737292 | 0.14 |
ENST00000254231.4 |
LIN28A |
lin-28 homolog A (C. elegans) |
| chr12_-_49245936 | 0.14 |
ENST00000308025.3 |
DDX23 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 23 |
| chr10_-_103603568 | 0.14 |
ENST00000356640.2 |
KCNIP2 |
Kv channel interacting protein 2 |
| chrX_+_138612889 | 0.14 |
ENST00000218099.2 ENST00000394090.2 |
F9 |
coagulation factor IX |
| chr1_+_152975488 | 0.14 |
ENST00000542696.1 |
SPRR3 |
small proline-rich protein 3 |
| chr12_+_20848486 | 0.14 |
ENST00000545102.1 |
SLCO1C1 |
solute carrier organic anion transporter family, member 1C1 |
| chr18_-_53257027 | 0.13 |
ENST00000568740.1 ENST00000564403.2 ENST00000537578.1 |
TCF4 |
transcription factor 4 |
| chr11_+_70244510 | 0.13 |
ENST00000346329.3 ENST00000301843.8 ENST00000376561.3 |
CTTN |
cortactin |
| chr4_+_129349188 | 0.13 |
ENST00000511497.1 |
RP11-420A23.1 |
RP11-420A23.1 |
| chr7_-_71912046 | 0.13 |
ENST00000395276.2 ENST00000431984.1 |
CALN1 |
calneuron 1 |
| chr11_+_55594695 | 0.13 |
ENST00000378397.1 |
OR5L2 |
olfactory receptor, family 5, subfamily L, member 2 |
| chr10_-_6622258 | 0.13 |
ENST00000263125.5 |
PRKCQ |
protein kinase C, theta |
| chr21_+_27011584 | 0.12 |
ENST00000400532.1 ENST00000480456.1 ENST00000312957.5 |
JAM2 |
junctional adhesion molecule 2 |
| chr21_+_27011899 | 0.11 |
ENST00000425221.2 |
JAM2 |
junctional adhesion molecule 2 |
| chr6_-_167797887 | 0.11 |
ENST00000476779.2 ENST00000460930.2 ENST00000397829.4 ENST00000366827.2 |
TCP10 |
t-complex 10 |
| chr11_-_104769141 | 0.11 |
ENST00000508062.1 ENST00000422698.2 |
CASP12 |
caspase 12 (gene/pseudogene) |
| chr12_-_55378452 | 0.11 |
ENST00000449076.1 |
TESPA1 |
thymocyte expressed, positive selection associated 1 |
| chr5_-_35195338 | 0.11 |
ENST00000509839.1 |
PRLR |
prolactin receptor |
| chrX_+_152907913 | 0.10 |
ENST00000370167.4 |
DUSP9 |
dual specificity phosphatase 9 |
| chr2_-_18770812 | 0.10 |
ENST00000359846.2 ENST00000304081.4 ENST00000600945.1 ENST00000532967.1 ENST00000444297.2 |
NT5C1B NT5C1B-RDH14 |
5'-nucleotidase, cytosolic IB NT5C1B-RDH14 readthrough |
| chr2_-_89597542 | 0.10 |
ENST00000465170.1 |
IGKV1-37 |
immunoglobulin kappa variable 1-37 (non-functional) |
| chr4_+_155484155 | 0.10 |
ENST00000509493.1 |
FGB |
fibrinogen beta chain |
| chr10_-_103603677 | 0.10 |
ENST00000358038.3 |
KCNIP2 |
Kv channel interacting protein 2 |
| chr12_+_19358228 | 0.10 |
ENST00000424268.1 ENST00000543806.1 |
PLEKHA5 |
pleckstrin homology domain containing, family A member 5 |
| chr12_-_6756609 | 0.10 |
ENST00000229243.2 |
ACRBP |
acrosin binding protein |
| chr11_-_104840093 | 0.10 |
ENST00000417440.2 ENST00000444739.2 |
CASP4 |
caspase 4, apoptosis-related cysteine peptidase |
| chr19_-_11039261 | 0.10 |
ENST00000590329.1 ENST00000587943.1 ENST00000585858.1 ENST00000586748.1 ENST00000586575.1 ENST00000253031.2 |
YIPF2 |
Yip1 domain family, member 2 |
| chr1_+_196743912 | 0.09 |
ENST00000367425.4 |
CFHR3 |
complement factor H-related 3 |
| chr8_-_19540266 | 0.09 |
ENST00000311540.4 |
CSGALNACT1 |
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr8_-_19540086 | 0.09 |
ENST00000332246.6 |
CSGALNACT1 |
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr11_-_5173599 | 0.09 |
ENST00000328942.1 |
OR52A1 |
olfactory receptor, family 52, subfamily A, member 1 |
| chr12_-_50790267 | 0.09 |
ENST00000327337.5 ENST00000543111.1 |
FAM186A |
family with sequence similarity 186, member A |
| chr8_-_124749609 | 0.09 |
ENST00000262219.6 ENST00000419625.1 |
ANXA13 |
annexin A13 |
| chr7_+_75511362 | 0.09 |
ENST00000428119.1 |
RHBDD2 |
rhomboid domain containing 2 |
| chr10_-_103603523 | 0.09 |
ENST00000370046.1 |
KCNIP2 |
Kv channel interacting protein 2 |
| chr11_+_73661364 | 0.09 |
ENST00000339764.1 |
DNAJB13 |
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr14_+_22771851 | 0.09 |
ENST00000390466.1 |
TRAV39 |
T cell receptor alpha variable 39 |
| chr6_+_88757507 | 0.09 |
ENST00000237201.1 |
SPACA1 |
sperm acrosome associated 1 |
| chr19_-_11039188 | 0.08 |
ENST00000588347.1 |
YIPF2 |
Yip1 domain family, member 2 |
| chr1_-_13390765 | 0.08 |
ENST00000357367.2 |
PRAMEF8 |
PRAME family member 8 |
| chr11_-_44972418 | 0.08 |
ENST00000525680.1 ENST00000528290.1 ENST00000530035.1 |
TP53I11 |
tumor protein p53 inducible protein 11 |
| chr17_-_8151353 | 0.08 |
ENST00000315684.8 |
CTC1 |
CTS telomere maintenance complex component 1 |
| chr19_+_56459198 | 0.08 |
ENST00000291971.3 ENST00000590542.1 |
NLRP8 |
NLR family, pyrin domain containing 8 |
| chr4_+_159131346 | 0.08 |
ENST00000508243.1 ENST00000296529.6 |
TMEM144 |
transmembrane protein 144 |
| chr6_-_137365402 | 0.08 |
ENST00000541547.1 |
IL20RA |
interleukin 20 receptor, alpha |
| chr6_-_29343068 | 0.08 |
ENST00000396806.3 |
OR12D3 |
olfactory receptor, family 12, subfamily D, member 3 |
| chr2_+_105050794 | 0.08 |
ENST00000429464.1 ENST00000414442.1 ENST00000447380.1 |
AC013402.2 |
long intergenic non-protein coding RNA 1102 |
| chr1_+_87595433 | 0.07 |
ENST00000469312.2 ENST00000490006.2 |
RP5-1052I5.1 |
long intergenic non-protein coding RNA 1140 |
| chr9_+_131644388 | 0.07 |
ENST00000372600.4 |
LRRC8A |
leucine rich repeat containing 8 family, member A |
| chr2_+_168043793 | 0.07 |
ENST00000409273.1 ENST00000409605.1 |
XIRP2 |
xin actin-binding repeat containing 2 |
| chr12_+_100750846 | 0.07 |
ENST00000323346.5 |
SLC17A8 |
solute carrier family 17 (vesicular glutamate transporter), member 8 |
| chr14_+_22675388 | 0.07 |
ENST00000390461.2 |
TRAV34 |
T cell receptor alpha variable 34 |
| chr17_-_56769382 | 0.07 |
ENST00000240361.8 ENST00000349033.5 ENST00000389934.3 |
TEX14 |
testis expressed 14 |
| chr2_+_89923550 | 0.07 |
ENST00000509129.1 |
IGKV1D-37 |
immunoglobulin kappa variable 1D-37 (non-functional) |
| chr2_+_219135115 | 0.07 |
ENST00000248451.3 ENST00000273077.4 |
PNKD |
paroxysmal nonkinesigenic dyskinesia |
| chr9_+_132427883 | 0.07 |
ENST00000372469.4 |
PRRX2 |
paired related homeobox 2 |
| chr9_+_130830451 | 0.07 |
ENST00000373068.2 ENST00000373069.5 |
SLC25A25 |
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25 |
| chr9_+_131644398 | 0.07 |
ENST00000372599.3 |
LRRC8A |
leucine rich repeat containing 8 family, member A |
| chr19_+_17862274 | 0.07 |
ENST00000596536.1 ENST00000593870.1 ENST00000598086.1 ENST00000598932.1 ENST00000595023.1 ENST00000594068.1 ENST00000596507.1 ENST00000595033.1 ENST00000597718.1 |
FCHO1 |
FCH domain only 1 |
| chr12_-_96429423 | 0.07 |
ENST00000228740.2 |
LTA4H |
leukotriene A4 hydrolase |
| chr22_+_22936998 | 0.07 |
ENST00000390303.2 |
IGLV3-32 |
immunoglobulin lambda variable 3-32 (non-functional) |
| chr5_+_140201183 | 0.07 |
ENST00000529619.1 ENST00000529859.1 ENST00000378126.3 |
PCDHA5 |
protocadherin alpha 5 |
| chr18_+_616672 | 0.07 |
ENST00000338387.7 |
CLUL1 |
clusterin-like 1 (retinal) |
| chr9_+_84603687 | 0.07 |
ENST00000344803.2 |
SPATA31D1 |
SPATA31 subfamily D, member 1 |
| chr1_-_47082495 | 0.07 |
ENST00000545730.1 ENST00000531769.1 ENST00000319928.3 |
MKNK1 MOB3C |
MAP kinase interacting serine/threonine kinase 1 MOB kinase activator 3C |
| chr6_-_42690312 | 0.07 |
ENST00000230381.5 |
PRPH2 |
peripherin 2 (retinal degeneration, slow) |
| chr12_-_10962767 | 0.07 |
ENST00000240691.2 |
TAS2R9 |
taste receptor, type 2, member 9 |
| chr20_-_1538319 | 0.07 |
ENST00000381621.1 |
SIRPD |
signal-regulatory protein delta |
| chr10_+_5005598 | 0.07 |
ENST00000442997.1 |
AKR1C1 |
aldo-keto reductase family 1, member C1 |
| chr21_-_34185944 | 0.07 |
ENST00000479548.1 |
C21orf62 |
chromosome 21 open reading frame 62 |
| chr7_-_48068643 | 0.07 |
ENST00000453192.2 |
SUN3 |
Sad1 and UNC84 domain containing 3 |
| chr4_+_80748619 | 0.06 |
ENST00000504263.1 |
PCAT4 |
prostate cancer associated transcript 4 (non-protein coding) |
| chr19_+_1026298 | 0.06 |
ENST00000263097.4 |
CNN2 |
calponin 2 |
| chr8_+_95835438 | 0.06 |
ENST00000521860.1 ENST00000519457.1 ENST00000519053.1 ENST00000523731.1 ENST00000447247.1 |
INTS8 |
integrator complex subunit 8 |
| chr12_+_81471816 | 0.06 |
ENST00000261206.3 |
ACSS3 |
acyl-CoA synthetase short-chain family member 3 |
| chr4_-_156298087 | 0.06 |
ENST00000311277.4 |
MAP9 |
microtubule-associated protein 9 |
| chr1_-_44818599 | 0.06 |
ENST00000537474.1 |
ERI3 |
ERI1 exoribonuclease family member 3 |
| chr3_-_129147432 | 0.06 |
ENST00000503957.1 ENST00000505956.1 ENST00000326085.3 |
EFCAB12 |
EF-hand calcium binding domain 12 |
| chr1_+_12976450 | 0.06 |
ENST00000361079.2 |
PRAMEF7 |
PRAME family member 7 |
| chr3_+_196669494 | 0.06 |
ENST00000602845.1 |
NCBP2-AS2 |
NCBP2 antisense RNA 2 (head to head) |
| chrX_+_8432871 | 0.06 |
ENST00000381032.1 ENST00000453306.1 ENST00000444481.1 |
VCX3B |
variable charge, X-linked 3B |
| chr13_-_41837620 | 0.06 |
ENST00000379477.1 ENST00000452359.1 ENST00000379480.4 ENST00000430347.2 |
MTRF1 |
mitochondrial translational release factor 1 |
| chr2_+_105953972 | 0.06 |
ENST00000410049.1 |
C2orf49 |
chromosome 2 open reading frame 49 |
| chr7_+_72349920 | 0.06 |
ENST00000395270.1 ENST00000446813.1 ENST00000257622.4 |
POM121 |
POM121 transmembrane nucleoporin |
| chr11_+_85339623 | 0.06 |
ENST00000358867.6 ENST00000534341.1 ENST00000393375.1 ENST00000531274.1 |
TMEM126B |
transmembrane protein 126B |
| chr6_-_31509714 | 0.06 |
ENST00000456662.1 ENST00000431908.1 ENST00000456976.1 ENST00000428450.1 ENST00000453105.2 ENST00000418897.1 ENST00000415382.2 ENST00000449074.2 ENST00000419020.1 ENST00000428098.1 |
DDX39B |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chr2_-_73053126 | 0.06 |
ENST00000272427.6 ENST00000410104.1 |
EXOC6B |
exocyst complex component 6B |
| chr7_-_48068671 | 0.06 |
ENST00000297325.4 |
SUN3 |
Sad1 and UNC84 domain containing 3 |
| chr11_-_111794446 | 0.06 |
ENST00000527950.1 |
CRYAB |
crystallin, alpha B |
| chr19_-_39226045 | 0.06 |
ENST00000597987.1 ENST00000595177.1 |
CAPN12 |
calpain 12 |
| chr14_-_95942173 | 0.06 |
ENST00000334258.5 ENST00000557275.1 ENST00000553340.1 |
SYNE3 |
spectrin repeat containing, nuclear envelope family member 3 |
| chr1_-_158656488 | 0.06 |
ENST00000368147.4 |
SPTA1 |
spectrin, alpha, erythrocytic 1 (elliptocytosis 2) |
| chr3_+_178253993 | 0.06 |
ENST00000420517.2 ENST00000452583.1 |
KCNMB2 |
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr19_+_1524072 | 0.06 |
ENST00000454744.2 |
PLK5 |
polo-like kinase 5 |
| chr11_+_36317830 | 0.06 |
ENST00000530639.1 |
PRR5L |
proline rich 5 like |
| chr15_-_43212996 | 0.05 |
ENST00000567840.1 |
TTBK2 |
tau tubulin kinase 2 |
| chr7_+_123241908 | 0.05 |
ENST00000434204.1 ENST00000437535.1 ENST00000451215.1 |
ASB15 |
ankyrin repeat and SOCS box containing 15 |
| chr7_+_76751926 | 0.05 |
ENST00000285871.4 ENST00000431197.1 |
CCDC146 |
coiled-coil domain containing 146 |
| chr5_-_59481406 | 0.05 |
ENST00000546160.1 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
| chr7_-_75452673 | 0.05 |
ENST00000416943.1 |
CCL24 |
chemokine (C-C motif) ligand 24 |
| chr4_+_119199904 | 0.05 |
ENST00000602483.1 ENST00000602819.1 |
SNHG8 |
small nucleolar RNA host gene 8 (non-protein coding) |
| chr14_-_24740709 | 0.05 |
ENST00000399409.3 ENST00000216840.6 |
RABGGTA |
Rab geranylgeranyltransferase, alpha subunit |
| chr19_+_15197791 | 0.05 |
ENST00000209540.2 |
OR1I1 |
olfactory receptor, family 1, subfamily I, member 1 |
| chr14_-_74296806 | 0.05 |
ENST00000555539.1 |
RP5-1021I20.2 |
RP5-1021I20.2 |
| chr17_-_6915646 | 0.05 |
ENST00000574377.1 ENST00000399541.2 ENST00000399540.2 ENST00000575727.1 ENST00000573939.1 |
AC027763.2 |
Uncharacterized protein |
| chr14_+_78266408 | 0.05 |
ENST00000238561.5 |
ADCK1 |
aarF domain containing kinase 1 |
| chr14_+_78266436 | 0.05 |
ENST00000557501.1 ENST00000341211.5 |
ADCK1 |
aarF domain containing kinase 1 |
| chr15_-_43212836 | 0.05 |
ENST00000566931.1 ENST00000564431.1 ENST00000567274.1 |
TTBK2 |
tau tubulin kinase 2 |
| chr21_-_33957805 | 0.05 |
ENST00000300258.3 ENST00000472557.1 |
TCP10L |
t-complex 10-like |
| chr2_-_180871780 | 0.05 |
ENST00000410053.3 ENST00000295749.6 ENST00000404136.2 |
CWC22 |
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
| chr12_-_5352315 | 0.05 |
ENST00000536518.1 |
RP11-319E16.1 |
RP11-319E16.1 |
| chr11_-_47788847 | 0.05 |
ENST00000263773.5 |
FNBP4 |
formin binding protein 4 |
| chr11_-_32452357 | 0.05 |
ENST00000379079.2 ENST00000530998.1 |
WT1 |
Wilms tumor 1 |
| chr7_-_48068699 | 0.05 |
ENST00000412142.1 ENST00000395572.2 |
SUN3 |
Sad1 and UNC84 domain containing 3 |
| chr16_+_29690358 | 0.05 |
ENST00000395384.4 ENST00000562473.1 |
QPRT |
quinolinate phosphoribosyltransferase |
| chr19_-_1848451 | 0.05 |
ENST00000170168.4 |
REXO1 |
REX1, RNA exonuclease 1 homolog (S. cerevisiae) |
| chr10_-_43762329 | 0.05 |
ENST00000395810.1 |
RASGEF1A |
RasGEF domain family, member 1A |
| chr15_+_75491213 | 0.05 |
ENST00000360639.2 |
C15orf39 |
chromosome 15 open reading frame 39 |
| chr17_+_37356528 | 0.05 |
ENST00000225430.4 |
RPL19 |
ribosomal protein L19 |
| chr17_-_34625719 | 0.05 |
ENST00000422211.2 ENST00000542124.1 |
CCL3L1 |
chemokine (C-C motif) ligand 3-like 1 |
| chr17_+_37356555 | 0.05 |
ENST00000579374.1 |
RPL19 |
ribosomal protein L19 |
| chr12_-_6798616 | 0.05 |
ENST00000355772.4 ENST00000417772.3 ENST00000396801.3 ENST00000396799.2 |
ZNF384 |
zinc finger protein 384 |
| chr15_+_59279851 | 0.05 |
ENST00000348370.4 ENST00000434298.1 ENST00000559160.1 |
RNF111 |
ring finger protein 111 |
| chr17_+_37356586 | 0.04 |
ENST00000579260.1 ENST00000582193.1 |
RPL19 |
ribosomal protein L19 |
| chr2_-_55496174 | 0.04 |
ENST00000417363.1 ENST00000412530.1 ENST00000394600.3 ENST00000366137.2 ENST00000420637.1 |
MTIF2 |
mitochondrial translational initiation factor 2 |
| chr17_+_4675175 | 0.04 |
ENST00000270560.3 |
TM4SF5 |
transmembrane 4 L six family member 5 |
| chr7_-_6866401 | 0.04 |
ENST00000316731.8 |
CCZ1B |
CCZ1 vacuolar protein trafficking and biogenesis associated homolog B (S. cerevisiae) |
| chr11_-_62559455 | 0.04 |
ENST00000528367.1 ENST00000525631.1 ENST00000307366.7 |
TMEM223 |
transmembrane protein 223 |
| chr4_-_76008706 | 0.04 |
ENST00000562355.1 ENST00000563602.1 |
RP11-44F21.5 |
RP11-44F21.5 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0060557 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.1 | 0.4 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 0.1 | 0.4 | GO:1900138 | negative regulation of phospholipase A2 activity(GO:1900138) |
| 0.1 | 0.6 | GO:2001268 | positive regulation of keratinocyte migration(GO:0051549) negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.1 | 0.3 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.1 | 0.3 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.3 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:0000354 | cis assembly of pre-catalytic spliceosome(GO:0000354) |
| 0.0 | 0.3 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.3 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.0 | 0.2 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.0 | 0.3 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.0 | 0.2 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.5 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:2000569 | T-helper 2 cell activation(GO:0035712) regulation of T-helper 2 cell activation(GO:2000569) positive regulation of T-helper 2 cell activation(GO:2000570) |
| 0.0 | 0.5 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.2 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.3 | GO:0016045 | detection of bacterium(GO:0016045) |
| 0.0 | 0.1 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.4 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.0 | 0.1 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.1 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.0 | 2.8 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.0 | 0.2 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.0 | 0.6 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.1 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.2 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.0 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.0 | 0.1 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 | 2.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.2 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.0 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.0 | 0.0 | GO:0035377 | transepithelial water transport(GO:0035377) positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.0 | 0.1 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.0 | GO:0021938 | ventral midline development(GO:0007418) smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.0 | 0.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.0 | 0.2 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.2 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.0 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 2.4 | GO:0005875 | microtubule associated complex(GO:0005875) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.1 | 0.3 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.1 | 0.7 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.3 | GO:0030305 | beta-glucuronidase activity(GO:0004566) heparanase activity(GO:0030305) |
| 0.1 | 0.2 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.1 | 0.5 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.3 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.3 | GO:0030883 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 0.3 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.3 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.3 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.4 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.0 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.0 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.0 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.0 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.0 | 0.0 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.0 | 0.1 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.2 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.1 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.9 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |