Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
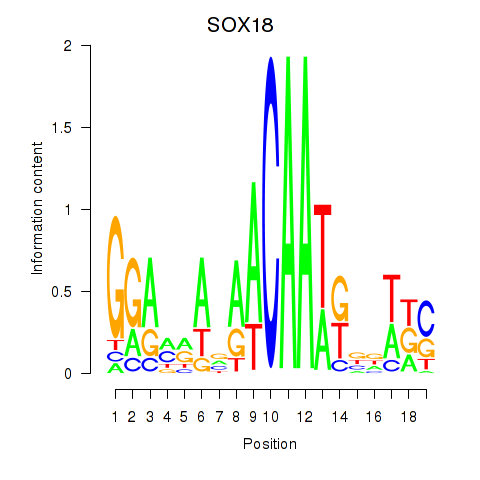
Results for SOX18
Z-value: 0.50
Transcription factors associated with SOX18
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX18
|
ENSG00000203883.5 | SOX18 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX18 | hg19_v2_chr20_-_62680984_62680999 | -0.36 | 3.7e-01 | Click! |
Activity profile of SOX18 motif
Sorted Z-values of SOX18 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX18
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_5710919 | 0.33 |
ENST00000379965.3 ENST00000425490.1 |
TRIM22 |
tripartite motif containing 22 |
| chr11_-_89223883 | 0.32 |
ENST00000528341.1 |
NOX4 |
NADPH oxidase 4 |
| chr11_-_89224139 | 0.32 |
ENST00000413594.2 |
NOX4 |
NADPH oxidase 4 |
| chr21_-_31744557 | 0.29 |
ENST00000399889.2 |
KRTAP13-2 |
keratin associated protein 13-2 |
| chr17_+_41158742 | 0.26 |
ENST00000415816.2 ENST00000438323.2 |
IFI35 |
interferon-induced protein 35 |
| chrX_-_154033661 | 0.23 |
ENST00000393531.1 |
MPP1 |
membrane protein, palmitoylated 1, 55kDa |
| chr4_-_57524061 | 0.21 |
ENST00000508121.1 |
HOPX |
HOP homeobox |
| chr7_+_141811539 | 0.21 |
ENST00000550469.2 ENST00000477922.3 |
RP11-1220K2.2 |
Putative inactive maltase-glucoamylase-like protein LOC93432 |
| chr2_-_202562774 | 0.21 |
ENST00000396886.3 ENST00000409143.1 |
MPP4 |
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4) |
| chr12_+_15475462 | 0.18 |
ENST00000543886.1 ENST00000348962.2 |
PTPRO |
protein tyrosine phosphatase, receptor type, O |
| chr13_-_33780133 | 0.16 |
ENST00000399365.3 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
| chr18_+_74240610 | 0.15 |
ENST00000578092.1 ENST00000578613.1 ENST00000583578.1 |
LINC00908 |
long intergenic non-protein coding RNA 908 |
| chr11_-_111749878 | 0.14 |
ENST00000260257.4 |
FDXACB1 |
ferredoxin-fold anticodon binding domain containing 1 |
| chr1_-_182361327 | 0.14 |
ENST00000331872.6 ENST00000311223.5 |
GLUL |
glutamate-ammonia ligase |
| chr4_+_109541772 | 0.14 |
ENST00000506397.1 ENST00000394668.2 |
RPL34 |
ribosomal protein L34 |
| chr11_-_111749767 | 0.14 |
ENST00000542429.1 |
FDXACB1 |
ferredoxin-fold anticodon binding domain containing 1 |
| chr1_-_182360918 | 0.14 |
ENST00000339526.4 |
GLUL |
glutamate-ammonia ligase |
| chr11_-_89224299 | 0.13 |
ENST00000343727.5 ENST00000531342.1 ENST00000375979.3 |
NOX4 |
NADPH oxidase 4 |
| chr20_-_44718538 | 0.13 |
ENST00000290231.6 ENST00000372291.3 |
NCOA5 |
nuclear receptor coactivator 5 |
| chr19_-_7040190 | 0.13 |
ENST00000381394.4 |
MBD3L4 |
methyl-CpG binding domain protein 3-like 4 |
| chr19_+_7030589 | 0.12 |
ENST00000329753.5 |
MBD3L5 |
methyl-CpG binding domain protein 3-like 5 |
| chrX_+_10031499 | 0.12 |
ENST00000454666.1 |
WWC3 |
WWC family member 3 |
| chr10_-_127505167 | 0.12 |
ENST00000368786.1 |
UROS |
uroporphyrinogen III synthase |
| chr8_-_13134045 | 0.12 |
ENST00000512044.2 |
DLC1 |
deleted in liver cancer 1 |
| chr12_-_15942503 | 0.12 |
ENST00000281172.5 |
EPS8 |
epidermal growth factor receptor pathway substrate 8 |
| chr6_-_152489484 | 0.11 |
ENST00000354674.4 ENST00000539504.1 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
| chr3_+_20081515 | 0.11 |
ENST00000263754.4 |
KAT2B |
K(lysine) acetyltransferase 2B |
| chr12_+_113344755 | 0.11 |
ENST00000550883.1 |
OAS1 |
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr2_-_158345462 | 0.11 |
ENST00000439355.1 ENST00000540637.1 |
CYTIP |
cytohesin 1 interacting protein |
| chr3_+_133465228 | 0.11 |
ENST00000482271.1 ENST00000264998.3 |
TF |
transferrin |
| chr14_-_71107921 | 0.11 |
ENST00000553982.1 ENST00000500016.1 |
CTD-2540L5.5 CTD-2540L5.6 |
CTD-2540L5.5 CTD-2540L5.6 |
| chr12_-_11002063 | 0.11 |
ENST00000544994.1 ENST00000228811.4 ENST00000540107.1 |
PRR4 |
proline rich 4 (lacrimal) |
| chr3_-_121379739 | 0.11 |
ENST00000428394.2 ENST00000314583.3 |
HCLS1 |
hematopoietic cell-specific Lyn substrate 1 |
| chr17_-_54893250 | 0.11 |
ENST00000397862.2 |
C17orf67 |
chromosome 17 open reading frame 67 |
| chr15_+_75628232 | 0.10 |
ENST00000267935.8 ENST00000567195.1 |
COMMD4 |
COMM domain containing 4 |
| chr6_+_89790459 | 0.10 |
ENST00000369472.1 |
PNRC1 |
proline-rich nuclear receptor coactivator 1 |
| chr19_+_40146534 | 0.10 |
ENST00000392051.3 |
LGALS16 |
lectin, galactoside-binding, soluble, 16 |
| chr7_-_6048650 | 0.10 |
ENST00000382321.4 ENST00000406569.3 |
PMS2 |
PMS2 postmeiotic segregation increased 2 (S. cerevisiae) |
| chr3_+_19189946 | 0.10 |
ENST00000328405.2 |
KCNH8 |
potassium voltage-gated channel, subfamily H (eag-related), member 8 |
| chr1_-_11918988 | 0.10 |
ENST00000376468.3 |
NPPB |
natriuretic peptide B |
| chr9_-_123812542 | 0.10 |
ENST00000223642.1 |
C5 |
complement component 5 |
| chr2_-_40739501 | 0.09 |
ENST00000403092.1 ENST00000402441.1 ENST00000448531.1 |
SLC8A1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr12_+_96252706 | 0.09 |
ENST00000266735.5 ENST00000553192.1 ENST00000552085.1 |
SNRPF |
small nuclear ribonucleoprotein polypeptide F |
| chr8_-_27469196 | 0.09 |
ENST00000546343.1 ENST00000560566.1 |
CLU |
clusterin |
| chr12_-_772901 | 0.09 |
ENST00000305108.4 |
NINJ2 |
ninjurin 2 |
| chr19_+_1249869 | 0.09 |
ENST00000591446.2 |
MIDN |
midnolin |
| chrX_+_47077632 | 0.09 |
ENST00000457458.2 |
CDK16 |
cyclin-dependent kinase 16 |
| chr22_+_37678424 | 0.09 |
ENST00000248901.6 |
CYTH4 |
cytohesin 4 |
| chr4_+_109541740 | 0.09 |
ENST00000394665.1 |
RPL34 |
ribosomal protein L34 |
| chr14_-_107219365 | 0.09 |
ENST00000424969.2 |
IGHV3-74 |
immunoglobulin heavy variable 3-74 |
| chr15_+_75628419 | 0.08 |
ENST00000567377.1 ENST00000562789.1 ENST00000568301.1 |
COMMD4 |
COMM domain containing 4 |
| chr22_-_22292934 | 0.08 |
ENST00000538191.1 ENST00000424647.1 ENST00000407142.1 |
PPM1F |
protein phosphatase, Mg2+/Mn2+ dependent, 1F |
| chr11_+_60102304 | 0.08 |
ENST00000300182.4 |
MS4A6E |
membrane-spanning 4-domains, subfamily A, member 6E |
| chr5_+_135468516 | 0.08 |
ENST00000507118.1 ENST00000511116.1 ENST00000545279.1 ENST00000545620.1 |
SMAD5 |
SMAD family member 5 |
| chr1_-_153517473 | 0.08 |
ENST00000368715.1 |
S100A4 |
S100 calcium binding protein A4 |
| chr12_-_11139511 | 0.08 |
ENST00000506868.1 |
TAS2R50 |
taste receptor, type 2, member 50 |
| chr3_+_57882024 | 0.08 |
ENST00000494088.1 |
SLMAP |
sarcolemma associated protein |
| chr14_+_22538811 | 0.08 |
ENST00000390450.3 |
TRAV22 |
T cell receptor alpha variable 22 |
| chr5_-_136834982 | 0.08 |
ENST00000510689.1 ENST00000394945.1 |
SPOCK1 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1 |
| chr8_-_27468842 | 0.08 |
ENST00000523500.1 |
CLU |
clusterin |
| chr2_-_100939195 | 0.07 |
ENST00000393437.3 |
LONRF2 |
LON peptidase N-terminal domain and ring finger 2 |
| chr9_-_99064386 | 0.07 |
ENST00000375262.2 |
HSD17B3 |
hydroxysteroid (17-beta) dehydrogenase 3 |
| chr15_+_75628394 | 0.07 |
ENST00000564815.1 ENST00000338995.6 |
COMMD4 |
COMM domain containing 4 |
| chr4_+_109541722 | 0.07 |
ENST00000394667.3 ENST00000502534.1 |
RPL34 |
ribosomal protein L34 |
| chr5_-_114598548 | 0.07 |
ENST00000379615.3 ENST00000419445.1 |
PGGT1B |
protein geranylgeranyltransferase type I, beta subunit |
| chr7_-_6048702 | 0.07 |
ENST00000265849.7 |
PMS2 |
PMS2 postmeiotic segregation increased 2 (S. cerevisiae) |
| chr10_-_8095412 | 0.07 |
ENST00000458727.1 ENST00000355358.1 |
RP11-379F12.3 GATA3-AS1 |
RP11-379F12.3 GATA3 antisense RNA 1 |
| chr2_+_234621551 | 0.07 |
ENST00000608381.1 ENST00000373414.3 |
UGT1A1 UGT1A5 |
UDP glucuronosyltransferase 1 family, polypeptide A8 UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr16_-_4896205 | 0.07 |
ENST00000589389.1 |
GLYR1 |
glyoxylate reductase 1 homolog (Arabidopsis) |
| chrX_+_52920336 | 0.07 |
ENST00000452154.2 |
FAM156B |
family with sequence similarity 156, member B |
| chr13_-_44735393 | 0.07 |
ENST00000400419.1 |
SMIM2 |
small integral membrane protein 2 |
| chr19_+_39971470 | 0.07 |
ENST00000607714.1 ENST00000599794.1 ENST00000597666.1 ENST00000601403.1 ENST00000602028.1 |
TIMM50 |
translocase of inner mitochondrial membrane 50 homolog (S. cerevisiae) |
| chr6_-_26235206 | 0.07 |
ENST00000244534.5 |
HIST1H1D |
histone cluster 1, H1d |
| chr14_+_24458123 | 0.07 |
ENST00000545240.1 ENST00000382755.4 |
DHRS4L2 |
dehydrogenase/reductase (SDR family) member 4 like 2 |
| chr6_+_89790490 | 0.07 |
ENST00000336032.3 |
PNRC1 |
proline-rich nuclear receptor coactivator 1 |
| chr3_-_197300194 | 0.07 |
ENST00000358186.2 ENST00000431056.1 |
BDH1 |
3-hydroxybutyrate dehydrogenase, type 1 |
| chr12_-_56321649 | 0.06 |
ENST00000454792.2 ENST00000408946.2 |
WIBG |
within bgcn homolog (Drosophila) |
| chr3_+_57881966 | 0.06 |
ENST00000495364.1 |
SLMAP |
sarcolemma associated protein |
| chr1_-_155880672 | 0.06 |
ENST00000609492.1 ENST00000368322.3 |
RIT1 |
Ras-like without CAAX 1 |
| chr18_-_5396271 | 0.06 |
ENST00000579951.1 |
EPB41L3 |
erythrocyte membrane protein band 4.1-like 3 |
| chr4_+_156588115 | 0.06 |
ENST00000455639.2 |
GUCY1A3 |
guanylate cyclase 1, soluble, alpha 3 |
| chrX_+_47078069 | 0.06 |
ENST00000357227.4 ENST00000519758.1 ENST00000520893.1 ENST00000517426.1 |
CDK16 |
cyclin-dependent kinase 16 |
| chr13_-_45992473 | 0.06 |
ENST00000539591.1 ENST00000519676.1 ENST00000519547.1 |
SLC25A30 |
solute carrier family 25, member 30 |
| chr19_+_39971505 | 0.06 |
ENST00000544017.1 |
TIMM50 |
translocase of inner mitochondrial membrane 50 homolog (S. cerevisiae) |
| chr16_-_28550320 | 0.06 |
ENST00000395641.2 |
NUPR1 |
nuclear protein, transcriptional regulator, 1 |
| chr9_-_74525847 | 0.06 |
ENST00000377041.2 |
ABHD17B |
abhydrolase domain containing 17B |
| chr5_+_31532373 | 0.06 |
ENST00000325366.9 ENST00000355907.3 ENST00000507818.2 |
C5orf22 |
chromosome 5 open reading frame 22 |
| chr18_+_3447572 | 0.06 |
ENST00000548489.2 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr12_-_68647281 | 0.06 |
ENST00000328087.4 ENST00000538666.1 |
IL22 |
interleukin 22 |
| chr19_+_48497901 | 0.06 |
ENST00000339841.2 |
ELSPBP1 |
epididymal sperm binding protein 1 |
| chr5_+_140767452 | 0.06 |
ENST00000519479.1 |
PCDHGB4 |
protocadherin gamma subfamily B, 4 |
| chr6_+_140175987 | 0.06 |
ENST00000414038.1 ENST00000431609.1 |
RP5-899B16.1 |
RP5-899B16.1 |
| chr3_+_160394940 | 0.06 |
ENST00000320767.2 |
ARL14 |
ADP-ribosylation factor-like 14 |
| chr12_+_32260085 | 0.06 |
ENST00000548411.1 ENST00000281474.5 ENST00000551086.1 |
BICD1 |
bicaudal D homolog 1 (Drosophila) |
| chr11_+_134201911 | 0.06 |
ENST00000389881.3 |
GLB1L2 |
galactosidase, beta 1-like 2 |
| chr19_+_8117636 | 0.05 |
ENST00000253451.4 ENST00000315626.4 |
CCL25 |
chemokine (C-C motif) ligand 25 |
| chr5_+_142125161 | 0.05 |
ENST00000432677.1 |
AC005592.1 |
AC005592.1 |
| chr12_-_110939870 | 0.05 |
ENST00000447578.2 ENST00000546588.1 ENST00000360579.7 ENST00000549970.1 ENST00000549578.1 |
VPS29 |
vacuolar protein sorting 29 homolog (S. cerevisiae) |
| chr1_-_45988542 | 0.05 |
ENST00000424390.1 |
PRDX1 |
peroxiredoxin 1 |
| chr15_+_44084040 | 0.05 |
ENST00000249786.4 |
SERF2 |
small EDRK-rich factor 2 |
| chr11_-_89224488 | 0.05 |
ENST00000534731.1 ENST00000527626.1 |
NOX4 |
NADPH oxidase 4 |
| chrX_-_49041242 | 0.05 |
ENST00000453382.1 ENST00000540849.1 ENST00000536904.1 ENST00000432913.1 |
PRICKLE3 |
prickle homolog 3 (Drosophila) |
| chr12_+_3000073 | 0.05 |
ENST00000397132.2 |
TULP3 |
tubby like protein 3 |
| chr12_+_8666126 | 0.05 |
ENST00000299665.2 |
CLEC4D |
C-type lectin domain family 4, member D |
| chr1_+_168756151 | 0.05 |
ENST00000420691.1 |
LINC00626 |
long intergenic non-protein coding RNA 626 |
| chr16_-_68034470 | 0.05 |
ENST00000412757.2 |
DPEP2 |
dipeptidase 2 |
| chr2_+_170590321 | 0.05 |
ENST00000392647.2 |
KLHL23 |
kelch-like family member 23 |
| chr19_+_36132631 | 0.05 |
ENST00000379026.2 ENST00000379023.4 ENST00000402764.2 ENST00000479824.1 |
ETV2 |
ets variant 2 |
| chr11_-_40315640 | 0.05 |
ENST00000278198.2 |
LRRC4C |
leucine rich repeat containing 4C |
| chr8_+_24151620 | 0.05 |
ENST00000437154.2 |
ADAM28 |
ADAM metallopeptidase domain 28 |
| chr5_+_140864649 | 0.05 |
ENST00000306593.1 |
PCDHGC4 |
protocadherin gamma subfamily C, 4 |
| chr4_+_156588249 | 0.05 |
ENST00000393832.3 |
GUCY1A3 |
guanylate cyclase 1, soluble, alpha 3 |
| chr7_-_93204033 | 0.05 |
ENST00000359558.2 ENST00000360249.4 ENST00000426151.1 |
CALCR |
calcitonin receptor |
| chr17_+_3118915 | 0.05 |
ENST00000304094.1 |
OR1A1 |
olfactory receptor, family 1, subfamily A, member 1 |
| chr1_+_206972215 | 0.05 |
ENST00000340758.2 |
IL19 |
interleukin 19 |
| chr11_-_89224508 | 0.05 |
ENST00000525196.1 |
NOX4 |
NADPH oxidase 4 |
| chr10_-_13390270 | 0.04 |
ENST00000378614.4 ENST00000545675.1 ENST00000327347.5 |
SEPHS1 |
selenophosphate synthetase 1 |
| chr5_+_147774275 | 0.04 |
ENST00000513826.1 |
FBXO38 |
F-box protein 38 |
| chr1_+_36348790 | 0.04 |
ENST00000373204.4 |
AGO1 |
argonaute RISC catalytic component 1 |
| chr5_-_179050066 | 0.04 |
ENST00000329433.6 ENST00000510411.1 |
HNRNPH1 |
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr5_+_54398463 | 0.04 |
ENST00000274306.6 |
GZMA |
granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3) |
| chr11_-_76155700 | 0.04 |
ENST00000572035.1 |
RP11-111M22.3 |
RP11-111M22.3 |
| chr7_+_151038850 | 0.04 |
ENST00000355851.4 ENST00000566856.1 ENST00000470229.1 |
NUB1 |
negative regulator of ubiquitin-like proteins 1 |
| chrX_+_36254051 | 0.04 |
ENST00000378657.4 |
CXorf30 |
chromosome X open reading frame 30 |
| chr1_+_206223941 | 0.04 |
ENST00000367126.4 |
AVPR1B |
arginine vasopressin receptor 1B |
| chr5_+_32585605 | 0.04 |
ENST00000265073.4 ENST00000515355.1 ENST00000502897.1 ENST00000510442.1 |
SUB1 |
SUB1 homolog (S. cerevisiae) |
| chr22_+_31488433 | 0.04 |
ENST00000455608.1 |
SMTN |
smoothelin |
| chr6_-_142409936 | 0.04 |
ENST00000258042.1 |
NMBR |
neuromedin B receptor |
| chr2_+_90259593 | 0.04 |
ENST00000471857.1 |
IGKV1D-8 |
immunoglobulin kappa variable 1D-8 |
| chr19_+_8117881 | 0.04 |
ENST00000390669.3 |
CCL25 |
chemokine (C-C motif) ligand 25 |
| chr1_+_43124087 | 0.04 |
ENST00000304979.3 ENST00000372550.1 ENST00000440068.1 |
PPIH |
peptidylprolyl isomerase H (cyclophilin H) |
| chr1_+_64669294 | 0.04 |
ENST00000371077.5 |
UBE2U |
ubiquitin-conjugating enzyme E2U (putative) |
| chr7_+_157129660 | 0.04 |
ENST00000429029.2 ENST00000262177.4 ENST00000417758.1 ENST00000452797.2 ENST00000443280.1 |
DNAJB6 |
DnaJ (Hsp40) homolog, subfamily B, member 6 |
| chr2_-_165811756 | 0.04 |
ENST00000409662.1 |
SLC38A11 |
solute carrier family 38, member 11 |
| chr9_-_128246769 | 0.04 |
ENST00000444226.1 |
MAPKAP1 |
mitogen-activated protein kinase associated protein 1 |
| chr2_-_211168332 | 0.04 |
ENST00000341685.4 |
MYL1 |
myosin, light chain 1, alkali; skeletal, fast |
| chr16_-_79015739 | 0.03 |
ENST00000594986.1 |
PIH1 |
HCG1979943; Uncharacterized protein |
| chr9_+_33240157 | 0.03 |
ENST00000379721.3 |
SPINK4 |
serine peptidase inhibitor, Kazal type 4 |
| chr1_+_161123536 | 0.03 |
ENST00000368003.5 |
UFC1 |
ubiquitin-fold modifier conjugating enzyme 1 |
| chr15_+_62359175 | 0.03 |
ENST00000355522.5 |
C2CD4A |
C2 calcium-dependent domain containing 4A |
| chr12_-_110883346 | 0.03 |
ENST00000547365.1 |
ARPC3 |
actin related protein 2/3 complex, subunit 3, 21kDa |
| chr12_-_49999389 | 0.03 |
ENST00000551047.1 ENST00000544141.1 |
FAM186B |
family with sequence similarity 186, member B |
| chr9_+_74526532 | 0.03 |
ENST00000486911.2 |
C9orf85 |
chromosome 9 open reading frame 85 |
| chr11_-_76155618 | 0.03 |
ENST00000530759.1 |
RP11-111M22.3 |
RP11-111M22.3 |
| chr3_+_140947563 | 0.03 |
ENST00000505013.1 |
ACPL2 |
acid phosphatase-like 2 |
| chr1_-_157746909 | 0.03 |
ENST00000392274.3 ENST00000361516.3 ENST00000368181.4 |
FCRL2 |
Fc receptor-like 2 |
| chr11_+_71238313 | 0.03 |
ENST00000398536.4 |
KRTAP5-7 |
keratin associated protein 5-7 |
| chr17_-_39296739 | 0.03 |
ENST00000345847.4 |
KRTAP4-6 |
keratin associated protein 4-6 |
| chr2_-_49381572 | 0.03 |
ENST00000454032.1 ENST00000304421.4 |
FSHR |
follicle stimulating hormone receptor |
| chr15_-_55541227 | 0.03 |
ENST00000566877.1 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr21_+_30671690 | 0.03 |
ENST00000399921.1 |
BACH1 |
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr10_-_4285923 | 0.03 |
ENST00000418372.1 ENST00000608792.1 |
LINC00702 |
long intergenic non-protein coding RNA 702 |
| chr16_+_33629600 | 0.03 |
ENST00000562905.2 |
IGHV3OR16-13 |
immunoglobulin heavy variable 3/OR16-13 (non-functional) |
| chr7_-_150329421 | 0.03 |
ENST00000493969.1 ENST00000328902.5 |
GIMAP6 |
GTPase, IMAP family member 6 |
| chr13_+_32313658 | 0.03 |
ENST00000380314.1 ENST00000298386.2 |
RXFP2 |
relaxin/insulin-like family peptide receptor 2 |
| chr19_-_57347415 | 0.03 |
ENST00000601070.1 |
ZIM2 |
zinc finger, imprinted 2 |
| chr1_-_155881156 | 0.03 |
ENST00000539040.1 ENST00000368323.3 |
RIT1 |
Ras-like without CAAX 1 |
| chr11_+_64949899 | 0.03 |
ENST00000531068.1 ENST00000527699.1 ENST00000533909.1 ENST00000527323.1 |
CAPN1 |
calpain 1, (mu/I) large subunit |
| chrX_+_9754461 | 0.03 |
ENST00000380913.3 |
SHROOM2 |
shroom family member 2 |
| chr19_+_57742431 | 0.03 |
ENST00000302804.7 |
AURKC |
aurora kinase C |
| chr11_+_4664650 | 0.03 |
ENST00000396952.5 |
OR51E1 |
olfactory receptor, family 51, subfamily E, member 1 |
| chr15_+_91473403 | 0.03 |
ENST00000394275.2 |
UNC45A |
unc-45 homolog A (C. elegans) |
| chr2_-_49381646 | 0.03 |
ENST00000346173.3 ENST00000406846.2 |
FSHR |
follicle stimulating hormone receptor |
| chr15_+_64386261 | 0.03 |
ENST00000560829.1 |
SNX1 |
sorting nexin 1 |
| chr1_-_206785789 | 0.03 |
ENST00000437518.1 ENST00000367114.3 |
EIF2D |
eukaryotic translation initiation factor 2D |
| chr2_-_47382414 | 0.03 |
ENST00000294947.2 |
C2orf61 |
chromosome 2 open reading frame 61 |
| chr16_-_787771 | 0.02 |
ENST00000568545.1 |
NARFL |
nuclear prelamin A recognition factor-like |
| chr1_+_225600404 | 0.02 |
ENST00000366845.2 |
AC092811.1 |
AC092811.1 |
| chr11_-_85397167 | 0.02 |
ENST00000316398.3 |
CCDC89 |
coiled-coil domain containing 89 |
| chr5_+_158690089 | 0.02 |
ENST00000296786.6 |
UBLCP1 |
ubiquitin-like domain containing CTD phosphatase 1 |
| chr6_-_75953484 | 0.02 |
ENST00000472311.2 ENST00000460985.1 ENST00000377978.3 ENST00000509698.1 ENST00000230459.4 ENST00000370089.2 |
COX7A2 |
cytochrome c oxidase subunit VIIa polypeptide 2 (liver) |
| chr2_-_153573965 | 0.02 |
ENST00000448428.1 |
PRPF40A |
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr2_+_79252822 | 0.02 |
ENST00000272324.5 |
REG3G |
regenerating islet-derived 3 gamma |
| chrX_+_100645812 | 0.02 |
ENST00000427805.2 ENST00000553110.3 ENST00000392994.3 ENST00000409338.1 ENST00000409170.3 |
RPL36A RPL36A-HNRNPH2 |
ribosomal protein L36a RPL36A-HNRNPH2 readthrough |
| chr7_+_12727250 | 0.02 |
ENST00000404894.1 |
ARL4A |
ADP-ribosylation factor-like 4A |
| chr13_-_53024725 | 0.02 |
ENST00000378060.4 |
VPS36 |
vacuolar protein sorting 36 homolog (S. cerevisiae) |
| chr2_+_79252804 | 0.02 |
ENST00000393897.2 |
REG3G |
regenerating islet-derived 3 gamma |
| chr1_-_206785898 | 0.02 |
ENST00000271764.2 |
EIF2D |
eukaryotic translation initiation factor 2D |
| chrX_+_2984874 | 0.02 |
ENST00000359361.2 |
ARSF |
arylsulfatase F |
| chr10_-_135187193 | 0.02 |
ENST00000368547.3 |
ECHS1 |
enoyl CoA hydratase, short chain, 1, mitochondrial |
| chrX_+_47053208 | 0.02 |
ENST00000442035.1 ENST00000457753.1 ENST00000335972.6 |
UBA1 |
ubiquitin-like modifier activating enzyme 1 |
| chr3_+_102153859 | 0.02 |
ENST00000306176.1 ENST00000466937.1 |
ZPLD1 |
zona pellucida-like domain containing 1 |
| chr9_-_74525658 | 0.02 |
ENST00000333421.6 |
ABHD17B |
abhydrolase domain containing 17B |
| chr11_-_118966167 | 0.02 |
ENST00000530167.1 |
H2AFX |
H2A histone family, member X |
| chr6_+_42883727 | 0.02 |
ENST00000304672.1 ENST00000441198.1 ENST00000446507.1 |
PTCRA |
pre T-cell antigen receptor alpha |
| chr19_+_44576296 | 0.02 |
ENST00000421176.3 |
ZNF284 |
zinc finger protein 284 |
| chr10_+_18429671 | 0.02 |
ENST00000282343.8 |
CACNB2 |
calcium channel, voltage-dependent, beta 2 subunit |
| chr3_+_186330712 | 0.02 |
ENST00000411641.2 ENST00000273784.5 |
AHSG |
alpha-2-HS-glycoprotein |
| chr20_+_59654146 | 0.02 |
ENST00000441660.1 |
RP5-827L5.1 |
RP5-827L5.1 |
| chr6_-_31125850 | 0.02 |
ENST00000507751.1 ENST00000448162.2 ENST00000502557.1 ENST00000503420.1 ENST00000507892.1 ENST00000507226.1 ENST00000513222.1 ENST00000503934.1 ENST00000396263.2 ENST00000508683.1 ENST00000428174.1 ENST00000448141.2 ENST00000507829.1 ENST00000455279.2 ENST00000376266.5 |
CCHCR1 |
coiled-coil alpha-helical rod protein 1 |
| chr8_-_37707356 | 0.02 |
ENST00000520601.1 ENST00000521170.1 ENST00000220659.6 |
BRF2 |
BRF2, RNA polymerase III transcription initiation factor 50 kDa subunit |
| chr1_-_12958101 | 0.02 |
ENST00000235347.4 |
PRAMEF10 |
PRAME family member 10 |
| chr4_-_114900831 | 0.02 |
ENST00000315366.7 |
ARSJ |
arylsulfatase family, member J |
| chr4_+_3443614 | 0.02 |
ENST00000382774.3 ENST00000511533.1 |
HGFAC |
HGF activator |
| chr11_-_118047376 | 0.02 |
ENST00000278947.5 |
SCN2B |
sodium channel, voltage-gated, type II, beta subunit |
| chr8_+_38644778 | 0.02 |
ENST00000276520.8 |
TACC1 |
transforming, acidic coiled-coil containing protein 1 |
| chr14_+_22217447 | 0.02 |
ENST00000390427.3 |
TRAV5 |
T cell receptor alpha variable 5 |
| chr16_+_66637777 | 0.02 |
ENST00000563672.1 ENST00000424011.2 |
CMTM3 |
CKLF-like MARVEL transmembrane domain containing 3 |
| chr10_+_82300575 | 0.02 |
ENST00000313455.4 |
SH2D4B |
SH2 domain containing 4B |
| chr2_+_105953972 | 0.01 |
ENST00000410049.1 |
C2orf49 |
chromosome 2 open reading frame 49 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.2 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.3 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.2 | GO:0061517 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.0 | 0.2 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.9 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.1 | GO:1900276 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.0 | 0.1 | GO:0052551 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.0 | 0.1 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 0.0 | 0.1 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.1 | GO:0097460 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.0 | 0.2 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.0 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.0 | 0.1 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.0 | 0.1 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:0060434 | bronchus morphogenesis(GO:0060434) |
| 0.0 | 0.1 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.3 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.9 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.0 | 0.2 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.1 | GO:0072517 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.3 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.0 | 0.1 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.2 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.9 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.3 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.1 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.0 | 0.1 | GO:0030395 | lactose binding(GO:0030395) |
| 0.0 | 0.2 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.0 | 0.1 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.0 | 0.1 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.0 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.1 | GO:0045518 | interleukin-22 receptor binding(GO:0045518) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.0 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.0 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |