Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
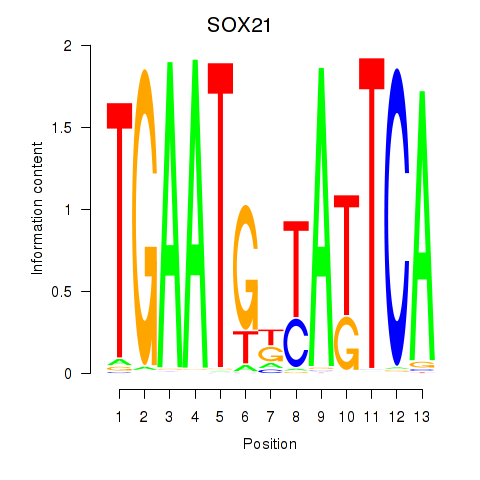
Results for SOX21
Z-value: 0.42
Transcription factors associated with SOX21
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX21
|
ENSG00000125285.4 | SOX21 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX21 | hg19_v2_chr13_-_95364389_95364389 | -0.73 | 4.0e-02 | Click! |
Activity profile of SOX21 motif
Sorted Z-values of SOX21 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX21
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_147286065 | 1.01 |
ENST00000318315.4 ENST00000515291.1 |
C5orf46 |
chromosome 5 open reading frame 46 |
| chr1_+_221051699 | 0.42 |
ENST00000366903.6 |
HLX |
H2.0-like homeobox |
| chr9_+_99690592 | 0.35 |
ENST00000354649.3 |
NUTM2G |
NUT family member 2G |
| chr2_+_102759199 | 0.32 |
ENST00000409288.1 ENST00000410023.1 |
IL1R1 |
interleukin 1 receptor, type I |
| chr3_+_141103634 | 0.31 |
ENST00000507722.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr5_-_95158644 | 0.30 |
ENST00000237858.6 |
GLRX |
glutaredoxin (thioltransferase) |
| chr5_-_95158375 | 0.29 |
ENST00000512469.2 ENST00000379979.4 ENST00000505427.1 ENST00000508780.1 |
GLRX |
glutaredoxin (thioltransferase) |
| chr17_-_8702667 | 0.27 |
ENST00000329805.4 |
MFSD6L |
major facilitator superfamily domain containing 6-like |
| chr5_-_61031495 | 0.25 |
ENST00000506550.1 ENST00000512882.2 |
CTD-2170G1.2 |
CTD-2170G1.2 |
| chr3_-_37034702 | 0.20 |
ENST00000322716.5 |
EPM2AIP1 |
EPM2A (laforin) interacting protein 1 |
| chr22_+_39353527 | 0.16 |
ENST00000249116.2 |
APOBEC3A |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3A |
| chr19_+_42381337 | 0.16 |
ENST00000597454.1 ENST00000444740.2 |
CD79A |
CD79a molecule, immunoglobulin-associated alpha |
| chr6_-_52926539 | 0.15 |
ENST00000350082.5 ENST00000356971.3 |
ICK |
intestinal cell (MAK-like) kinase |
| chr5_+_134181755 | 0.14 |
ENST00000504727.1 ENST00000435259.2 ENST00000508791.1 |
C5orf24 |
chromosome 5 open reading frame 24 |
| chr7_+_120702819 | 0.13 |
ENST00000423795.1 |
CPED1 |
cadherin-like and PC-esterase domain containing 1 |
| chr12_-_10588539 | 0.13 |
ENST00000381902.2 ENST00000381901.1 ENST00000539033.1 |
KLRC2 NKG2-E |
killer cell lectin-like receptor subfamily C, member 2 Uncharacterized protein |
| chr7_-_73133959 | 0.12 |
ENST00000395155.3 ENST00000395154.3 ENST00000222812.3 ENST00000395156.3 |
STX1A |
syntaxin 1A (brain) |
| chr1_-_112032284 | 0.11 |
ENST00000414219.1 |
ADORA3 |
adenosine A3 receptor |
| chr10_-_13276329 | 0.11 |
ENST00000378681.3 ENST00000463405.2 |
UCMA |
upper zone of growth plate and cartilage matrix associated |
| chr6_+_131894284 | 0.10 |
ENST00000368087.3 ENST00000356962.2 |
ARG1 |
arginase 1 |
| chr3_+_38537763 | 0.10 |
ENST00000287675.5 ENST00000358249.2 ENST00000422077.2 |
EXOG |
endo/exonuclease (5'-3'), endonuclease G-like |
| chr3_+_38537960 | 0.09 |
ENST00000453767.1 |
EXOG |
endo/exonuclease (5'-3'), endonuclease G-like |
| chrX_+_52927576 | 0.08 |
ENST00000416841.2 |
FAM156B |
family with sequence similarity 156, member B |
| chr4_-_39034542 | 0.08 |
ENST00000344606.6 |
TMEM156 |
transmembrane protein 156 |
| chr4_+_81951957 | 0.08 |
ENST00000282701.2 |
BMP3 |
bone morphogenetic protein 3 |
| chrX_+_36053908 | 0.08 |
ENST00000378660.2 |
CHDC2 |
calponin homology domain containing 2 |
| chr12_-_1058849 | 0.07 |
ENST00000358495.3 |
RAD52 |
RAD52 homolog (S. cerevisiae) |
| chr17_-_71410794 | 0.07 |
ENST00000424778.1 |
SDK2 |
sidekick cell adhesion molecule 2 |
| chr3_+_37035263 | 0.07 |
ENST00000458205.2 ENST00000539477.1 |
MLH1 |
mutL homolog 1 |
| chrX_-_71792477 | 0.06 |
ENST00000421523.1 ENST00000415409.1 ENST00000373559.4 ENST00000373556.3 ENST00000373560.2 ENST00000373583.1 ENST00000429103.2 ENST00000373571.1 ENST00000373554.1 |
HDAC8 |
histone deacetylase 8 |
| chr3_-_186288097 | 0.06 |
ENST00000446782.1 |
TBCCD1 |
TBCC domain containing 1 |
| chr3_+_155838337 | 0.06 |
ENST00000490337.1 ENST00000389636.5 |
KCNAB1 |
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr17_+_66245341 | 0.06 |
ENST00000577985.1 |
AMZ2 |
archaelysin family metallopeptidase 2 |
| chr12_-_7261772 | 0.06 |
ENST00000545280.1 ENST00000543933.1 ENST00000545337.1 ENST00000544702.1 ENST00000266542.4 |
C1RL |
complement component 1, r subcomponent-like |
| chr3_-_164875850 | 0.06 |
ENST00000472120.1 |
RP11-747D18.1 |
RP11-747D18.1 |
| chr19_-_21950332 | 0.06 |
ENST00000598026.1 |
ZNF100 |
zinc finger protein 100 |
| chr11_+_5410607 | 0.06 |
ENST00000328611.3 |
OR51M1 |
olfactory receptor, family 51, subfamily M, member 1 |
| chr11_-_62474803 | 0.05 |
ENST00000533982.1 ENST00000360796.5 |
BSCL2 |
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr19_+_42381173 | 0.05 |
ENST00000221972.3 |
CD79A |
CD79a molecule, immunoglobulin-associated alpha |
| chr10_-_17243579 | 0.05 |
ENST00000525762.1 ENST00000412821.3 ENST00000351358.4 ENST00000377766.5 ENST00000358282.7 ENST00000488990.1 ENST00000377799.3 |
TRDMT1 |
tRNA aspartic acid methyltransferase 1 |
| chr18_+_29769978 | 0.05 |
ENST00000269202.6 ENST00000581447.1 |
MEP1B |
meprin A, beta |
| chr3_+_37035289 | 0.05 |
ENST00000455445.2 ENST00000441265.1 ENST00000435176.1 ENST00000429117.1 ENST00000536378.1 |
MLH1 |
mutL homolog 1 |
| chr14_-_102771462 | 0.05 |
ENST00000522874.1 |
MOK |
MOK protein kinase |
| chr5_+_175875349 | 0.04 |
ENST00000261942.6 |
FAF2 |
Fas associated factor family member 2 |
| chr1_+_158901329 | 0.04 |
ENST00000368140.1 ENST00000368138.3 ENST00000392254.2 ENST00000392252.3 ENST00000368135.4 |
PYHIN1 |
pyrin and HIN domain family, member 1 |
| chrX_+_2984874 | 0.04 |
ENST00000359361.2 |
ARSF |
arylsulfatase F |
| chr14_-_92588013 | 0.04 |
ENST00000553514.1 ENST00000605997.1 |
NDUFB1 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 1, 7kDa |
| chr11_+_114168085 | 0.04 |
ENST00000541754.1 |
NNMT |
nicotinamide N-methyltransferase |
| chr8_-_19459993 | 0.04 |
ENST00000454498.2 ENST00000520003.1 |
CSGALNACT1 |
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr12_-_1058685 | 0.03 |
ENST00000397230.2 ENST00000542785.1 ENST00000544742.1 ENST00000536177.1 ENST00000539046.1 ENST00000541619.1 |
RAD52 |
RAD52 homolog (S. cerevisiae) |
| chr16_-_15149828 | 0.03 |
ENST00000566419.1 ENST00000568320.1 |
NTAN1 |
N-terminal asparagine amidase |
| chr5_-_179045199 | 0.03 |
ENST00000523921.1 |
HNRNPH1 |
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr9_-_70465758 | 0.03 |
ENST00000489273.1 |
CBWD5 |
COBW domain containing 5 |
| chr7_-_14942944 | 0.03 |
ENST00000403951.2 |
DGKB |
diacylglycerol kinase, beta 90kDa |
| chr7_-_86849883 | 0.03 |
ENST00000433078.1 |
TMEM243 |
transmembrane protein 243, mitochondrial |
| chr8_-_94029882 | 0.03 |
ENST00000520686.1 |
TRIQK |
triple QxxK/R motif containing |
| chr20_+_43343886 | 0.03 |
ENST00000190983.4 |
WISP2 |
WNT1 inducible signaling pathway protein 2 |
| chr6_+_140175987 | 0.03 |
ENST00000414038.1 ENST00000431609.1 |
RP5-899B16.1 |
RP5-899B16.1 |
| chr4_-_177190364 | 0.03 |
ENST00000296525.3 |
ASB5 |
ankyrin repeat and SOCS box containing 5 |
| chr10_-_12084770 | 0.03 |
ENST00000357604.5 |
UPF2 |
UPF2 regulator of nonsense transcripts homolog (yeast) |
| chrX_-_39186610 | 0.02 |
ENST00000429281.1 ENST00000448597.1 |
RP11-265P11.2 |
RP11-265P11.2 |
| chr11_-_57479673 | 0.02 |
ENST00000337672.2 ENST00000431606.2 |
MED19 |
mediator complex subunit 19 |
| chr1_+_199996733 | 0.02 |
ENST00000236914.3 |
NR5A2 |
nuclear receptor subfamily 5, group A, member 2 |
| chr3_+_37034823 | 0.02 |
ENST00000231790.2 ENST00000456676.2 |
MLH1 |
mutL homolog 1 |
| chr8_-_143961236 | 0.02 |
ENST00000377675.3 ENST00000517471.1 ENST00000292427.4 |
CYP11B1 |
cytochrome P450, family 11, subfamily B, polypeptide 1 |
| chr5_+_140579162 | 0.02 |
ENST00000536699.1 ENST00000354757.3 |
PCDHB11 |
protocadherin beta 11 |
| chr14_-_92588246 | 0.02 |
ENST00000329559.3 |
NDUFB1 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 1, 7kDa |
| chr22_-_40929812 | 0.02 |
ENST00000422851.1 |
MKL1 |
megakaryoblastic leukemia (translocation) 1 |
| chr12_+_123011776 | 0.01 |
ENST00000450485.2 ENST00000333479.7 |
KNTC1 |
kinetochore associated 1 |
| chr1_+_151171012 | 0.01 |
ENST00000349792.5 ENST00000409426.1 ENST00000441902.2 ENST00000368890.4 ENST00000424999.1 ENST00000368888.4 |
PIP5K1A |
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha |
| chr3_-_155572164 | 0.01 |
ENST00000392845.3 ENST00000359479.3 |
SLC33A1 |
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr8_+_67687413 | 0.01 |
ENST00000521960.1 ENST00000522398.1 ENST00000522629.1 ENST00000520976.1 ENST00000396596.1 |
SGK3 |
serum/glucocorticoid regulated kinase family, member 3 |
| chr14_+_21498360 | 0.01 |
ENST00000321760.6 ENST00000460647.2 ENST00000530140.2 ENST00000472458.1 |
TPPP2 |
tubulin polymerization-promoting protein family member 2 |
| chr11_-_8285405 | 0.01 |
ENST00000335790.3 ENST00000534484.1 |
LMO1 |
LIM domain only 1 (rhombotin 1) |
| chr9_+_99691286 | 0.01 |
ENST00000372322.3 |
NUTM2G |
NUT family member 2G |
| chr4_-_83931862 | 0.01 |
ENST00000506560.1 ENST00000442461.2 ENST00000446851.2 ENST00000340417.3 |
LIN54 |
lin-54 homolog (C. elegans) |
| chr12_-_10324716 | 0.01 |
ENST00000545927.1 ENST00000432556.2 ENST00000309539.3 ENST00000544577.1 |
OLR1 |
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr6_-_112575758 | 0.01 |
ENST00000431543.2 ENST00000453937.2 ENST00000368638.4 ENST00000389463.4 |
LAMA4 |
laminin, alpha 4 |
| chrY_+_16634483 | 0.01 |
ENST00000382872.1 |
NLGN4Y |
neuroligin 4, Y-linked |
| chr12_+_69742121 | 0.00 |
ENST00000261267.2 ENST00000549690.1 ENST00000548839.1 |
LYZ |
lysozyme |
| chr19_+_16308659 | 0.00 |
ENST00000590263.1 ENST00000590756.1 ENST00000541844.1 |
AP1M1 |
adaptor-related protein complex 1, mu 1 subunit |
| chr20_+_56964253 | 0.00 |
ENST00000395802.3 |
VAPB |
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr2_+_217363559 | 0.00 |
ENST00000600880.1 ENST00000446558.1 |
RPL37A |
ribosomal protein L37a |
| chr19_-_2783363 | 0.00 |
ENST00000221566.2 |
SGTA |
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.1 | 0.4 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.1 | GO:0051311 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 0.0 | 0.2 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.0 | 0.6 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.0 | 0.1 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.2 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.0 | 0.1 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.2 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 | 0.1 | GO:0043091 | L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.0 | 0.1 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 0.0 | 0.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) maintenance of Golgi location(GO:0051684) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005715 | late recombination nodule(GO:0005715) |
| 0.0 | 0.2 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.0 | 0.6 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.2 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.0 | 0.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.1 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 0.0 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |