Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
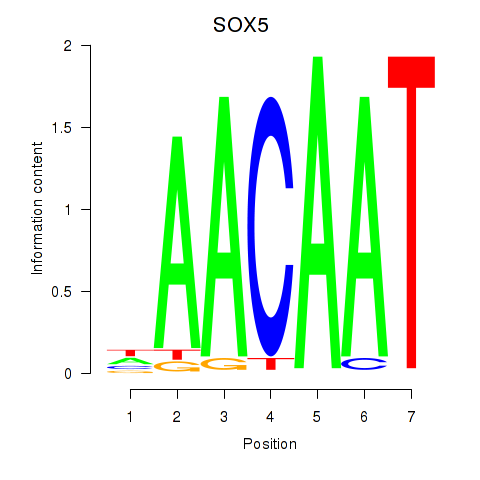
Results for SOX5
Z-value: 0.81
Transcription factors associated with SOX5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX5
|
ENSG00000134532.11 | SOX5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX5 | hg19_v2_chr12_-_24103954_24103972 | -0.78 | 2.4e-02 | Click! |
Activity profile of SOX5 motif
Sorted Z-values of SOX5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_92414055 | 2.10 |
ENST00000342058.4 |
FBLN5 |
fibulin 5 |
| chr14_-_92413727 | 1.74 |
ENST00000267620.10 |
FBLN5 |
fibulin 5 |
| chr14_-_92413353 | 1.22 |
ENST00000556154.1 |
FBLN5 |
fibulin 5 |
| chr1_+_163038565 | 0.93 |
ENST00000421743.2 |
RGS4 |
regulator of G-protein signaling 4 |
| chr3_-_18466026 | 0.75 |
ENST00000417717.2 |
SATB1 |
SATB homeobox 1 |
| chr14_-_22005018 | 0.65 |
ENST00000546363.1 |
SALL2 |
spalt-like transcription factor 2 |
| chr5_-_158526693 | 0.57 |
ENST00000380654.4 |
EBF1 |
early B-cell factor 1 |
| chr6_+_89790459 | 0.56 |
ENST00000369472.1 |
PNRC1 |
proline-rich nuclear receptor coactivator 1 |
| chr5_-_158526756 | 0.55 |
ENST00000313708.6 ENST00000517373.1 |
EBF1 |
early B-cell factor 1 |
| chr4_-_2264015 | 0.55 |
ENST00000337190.2 |
MXD4 |
MAX dimerization protein 4 |
| chr14_-_89883412 | 0.52 |
ENST00000557258.1 |
FOXN3 |
forkhead box N3 |
| chr3_-_114790179 | 0.49 |
ENST00000462705.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr12_+_53443680 | 0.47 |
ENST00000314250.6 ENST00000451358.1 |
TENC1 |
tensin like C1 domain containing phosphatase (tensin 2) |
| chr12_+_53443963 | 0.47 |
ENST00000546602.1 ENST00000552570.1 ENST00000549700.1 |
TENC1 |
tensin like C1 domain containing phosphatase (tensin 2) |
| chrX_-_106959631 | 0.45 |
ENST00000486554.1 ENST00000372390.4 |
TSC22D3 |
TSC22 domain family, member 3 |
| chrX_-_142722897 | 0.43 |
ENST00000338017.4 |
SLITRK4 |
SLIT and NTRK-like family, member 4 |
| chr6_+_89790490 | 0.42 |
ENST00000336032.3 |
PNRC1 |
proline-rich nuclear receptor coactivator 1 |
| chr8_-_93115445 | 0.41 |
ENST00000523629.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr20_-_45985172 | 0.41 |
ENST00000536340.1 |
ZMYND8 |
zinc finger, MYND-type containing 8 |
| chr5_+_67588391 | 0.38 |
ENST00000523872.1 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chrX_+_135251835 | 0.38 |
ENST00000456445.1 |
FHL1 |
four and a half LIM domains 1 |
| chr10_+_99079008 | 0.35 |
ENST00000371021.3 |
FRAT1 |
frequently rearranged in advanced T-cell lymphomas |
| chr1_+_164528866 | 0.35 |
ENST00000420696.2 |
PBX1 |
pre-B-cell leukemia homeobox 1 |
| chr1_-_86043921 | 0.33 |
ENST00000535924.2 |
DDAH1 |
dimethylarginine dimethylaminohydrolase 1 |
| chr9_-_123239632 | 0.33 |
ENST00000416449.1 |
CDK5RAP2 |
CDK5 regulatory subunit associated protein 2 |
| chrX_+_135251783 | 0.32 |
ENST00000394153.2 |
FHL1 |
four and a half LIM domains 1 |
| chr14_-_73493784 | 0.31 |
ENST00000553891.1 |
ZFYVE1 |
zinc finger, FYVE domain containing 1 |
| chr3_+_154797428 | 0.31 |
ENST00000460393.1 |
MME |
membrane metallo-endopeptidase |
| chr3_-_99594948 | 0.31 |
ENST00000471562.1 ENST00000495625.2 |
FILIP1L |
filamin A interacting protein 1-like |
| chr1_+_33722080 | 0.30 |
ENST00000483388.1 ENST00000539719.1 |
ZNF362 |
zinc finger protein 362 |
| chr9_+_118916082 | 0.29 |
ENST00000328252.3 |
PAPPA |
pregnancy-associated plasma protein A, pappalysin 1 |
| chr1_+_164529004 | 0.29 |
ENST00000559240.1 ENST00000367897.1 ENST00000540236.1 ENST00000401534.1 |
PBX1 |
pre-B-cell leukemia homeobox 1 |
| chr7_-_27142290 | 0.28 |
ENST00000222718.5 |
HOXA2 |
homeobox A2 |
| chrX_+_135252050 | 0.28 |
ENST00000449474.1 ENST00000345434.3 |
FHL1 |
four and a half LIM domains 1 |
| chr4_-_186732048 | 0.28 |
ENST00000448662.2 ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr11_-_82708435 | 0.28 |
ENST00000525117.1 ENST00000532548.1 |
RAB30 |
RAB30, member RAS oncogene family |
| chr1_-_232651312 | 0.28 |
ENST00000262861.4 |
SIPA1L2 |
signal-induced proliferation-associated 1 like 2 |
| chr6_-_152957944 | 0.28 |
ENST00000423061.1 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
| chrX_-_64196376 | 0.27 |
ENST00000447788.2 |
ZC4H2 |
zinc finger, C4H2 domain containing |
| chr14_-_73493825 | 0.27 |
ENST00000318876.5 ENST00000556143.1 |
ZFYVE1 |
zinc finger, FYVE domain containing 1 |
| chr3_-_114343039 | 0.27 |
ENST00000481632.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr7_-_47621736 | 0.27 |
ENST00000311160.9 |
TNS3 |
tensin 3 |
| chr2_-_200323414 | 0.27 |
ENST00000443023.1 |
SATB2 |
SATB homeobox 2 |
| chr10_+_35416090 | 0.27 |
ENST00000354759.3 |
CREM |
cAMP responsive element modulator |
| chr11_+_19799327 | 0.26 |
ENST00000540292.1 |
NAV2 |
neuron navigator 2 |
| chr20_-_45985414 | 0.26 |
ENST00000461685.1 ENST00000372023.3 ENST00000540497.1 ENST00000435836.1 ENST00000471951.2 ENST00000352431.2 ENST00000396281.4 ENST00000355972.4 ENST00000360911.3 |
ZMYND8 |
zinc finger, MYND-type containing 8 |
| chr11_+_19798964 | 0.26 |
ENST00000527559.2 |
NAV2 |
neuron navigator 2 |
| chr20_-_45985464 | 0.26 |
ENST00000458360.2 ENST00000262975.4 |
ZMYND8 |
zinc finger, MYND-type containing 8 |
| chr3_-_48754599 | 0.26 |
ENST00000413654.1 ENST00000454335.1 ENST00000440424.1 ENST00000449610.1 ENST00000443964.1 ENST00000417896.1 ENST00000413298.1 ENST00000449563.1 ENST00000443853.1 ENST00000437427.1 ENST00000446860.1 ENST00000412850.1 ENST00000424035.1 ENST00000340879.4 ENST00000431721.2 ENST00000434860.1 ENST00000328631.5 ENST00000432678.2 |
IP6K2 |
inositol hexakisphosphate kinase 2 |
| chr10_+_35415719 | 0.26 |
ENST00000474362.1 ENST00000374721.3 |
CREM |
cAMP responsive element modulator |
| chr1_-_156217822 | 0.25 |
ENST00000368270.1 |
PAQR6 |
progestin and adipoQ receptor family member VI |
| chr8_-_72274467 | 0.25 |
ENST00000340726.3 |
EYA1 |
eyes absent homolog 1 (Drosophila) |
| chr17_+_67498538 | 0.25 |
ENST00000589647.1 |
MAP2K6 |
mitogen-activated protein kinase kinase 6 |
| chr5_+_138677515 | 0.24 |
ENST00000265192.4 ENST00000511706.1 |
PAIP2 |
poly(A) binding protein interacting protein 2 |
| chr20_+_17207665 | 0.24 |
ENST00000536609.1 |
PCSK2 |
proprotein convertase subtilisin/kexin type 2 |
| chr1_-_156217829 | 0.24 |
ENST00000356983.2 ENST00000335852.1 ENST00000340183.5 ENST00000540423.1 |
PAQR6 |
progestin and adipoQ receptor family member VI |
| chr1_-_156217875 | 0.23 |
ENST00000292291.5 |
PAQR6 |
progestin and adipoQ receptor family member VI |
| chr15_-_52944231 | 0.23 |
ENST00000546305.2 |
FAM214A |
family with sequence similarity 214, member A |
| chr22_+_38093005 | 0.23 |
ENST00000406386.3 |
TRIOBP |
TRIO and F-actin binding protein |
| chr5_+_173316341 | 0.23 |
ENST00000520867.1 ENST00000334035.5 |
CPEB4 |
cytoplasmic polyadenylation element binding protein 4 |
| chr20_-_45981138 | 0.22 |
ENST00000446994.2 |
ZMYND8 |
zinc finger, MYND-type containing 8 |
| chr6_+_119215308 | 0.22 |
ENST00000229595.5 |
ASF1A |
anti-silencing function 1A histone chaperone |
| chr3_-_99595037 | 0.21 |
ENST00000383694.2 |
FILIP1L |
filamin A interacting protein 1-like |
| chr18_+_3451584 | 0.21 |
ENST00000551541.1 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr9_-_73483958 | 0.21 |
ENST00000377101.1 ENST00000377106.1 ENST00000360823.2 ENST00000377105.1 |
TRPM3 |
transient receptor potential cation channel, subfamily M, member 3 |
| chr11_-_82708519 | 0.21 |
ENST00000534301.1 |
RAB30 |
RAB30, member RAS oncogene family |
| chr2_-_133427767 | 0.21 |
ENST00000397463.2 |
LYPD1 |
LY6/PLAUR domain containing 1 |
| chr2_-_192711968 | 0.21 |
ENST00000304141.4 |
SDPR |
serum deprivation response |
| chrX_-_100872911 | 0.20 |
ENST00000361910.4 ENST00000539247.1 ENST00000538627.1 |
ARMCX6 |
armadillo repeat containing, X-linked 6 |
| chr5_-_61031495 | 0.20 |
ENST00000506550.1 ENST00000512882.2 |
CTD-2170G1.2 |
CTD-2170G1.2 |
| chr6_+_155537771 | 0.20 |
ENST00000275246.7 |
TIAM2 |
T-cell lymphoma invasion and metastasis 2 |
| chr9_+_82188077 | 0.20 |
ENST00000425506.1 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr1_+_205682497 | 0.19 |
ENST00000598338.1 |
AC119673.1 |
AC119673.1 |
| chr18_+_3451646 | 0.19 |
ENST00000345133.5 ENST00000330513.5 ENST00000549546.1 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr10_+_35415978 | 0.19 |
ENST00000429130.3 ENST00000469949.2 ENST00000460270.1 |
CREM |
cAMP responsive element modulator |
| chr12_-_92536433 | 0.19 |
ENST00000551563.2 ENST00000546975.1 ENST00000549802.1 |
C12orf79 |
chromosome 12 open reading frame 79 |
| chr20_-_18477862 | 0.19 |
ENST00000337227.4 |
RBBP9 |
retinoblastoma binding protein 9 |
| chr1_-_36906474 | 0.19 |
ENST00000433045.2 |
OSCP1 |
organic solute carrier partner 1 |
| chr10_+_63661053 | 0.19 |
ENST00000279873.7 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
| chr10_-_32667660 | 0.18 |
ENST00000375110.2 |
EPC1 |
enhancer of polycomb homolog 1 (Drosophila) |
| chr16_+_86612112 | 0.18 |
ENST00000320241.3 |
FOXL1 |
forkhead box L1 |
| chr19_+_19626531 | 0.18 |
ENST00000507754.4 |
NDUFA13 |
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 13 |
| chr4_+_110749143 | 0.18 |
ENST00000317735.4 |
RRH |
retinal pigment epithelium-derived rhodopsin homolog |
| chr10_+_104535994 | 0.18 |
ENST00000369889.4 |
WBP1L |
WW domain binding protein 1-like |
| chr3_+_141105705 | 0.18 |
ENST00000513258.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr7_+_134576317 | 0.18 |
ENST00000424922.1 ENST00000495522.1 |
CALD1 |
caldesmon 1 |
| chr16_-_73093597 | 0.17 |
ENST00000397992.5 |
ZFHX3 |
zinc finger homeobox 3 |
| chr6_+_126240442 | 0.17 |
ENST00000448104.1 ENST00000438495.1 ENST00000444128.1 |
NCOA7 |
nuclear receptor coactivator 7 |
| chr1_+_61547405 | 0.17 |
ENST00000371189.4 |
NFIA |
nuclear factor I/A |
| chr3_-_57233966 | 0.17 |
ENST00000473921.1 ENST00000295934.3 |
HESX1 |
HESX homeobox 1 |
| chr14_+_77228532 | 0.17 |
ENST00000167106.4 ENST00000554237.1 |
VASH1 |
vasohibin 1 |
| chr13_-_74708372 | 0.17 |
ENST00000377666.4 |
KLF12 |
Kruppel-like factor 12 |
| chr9_-_15510989 | 0.17 |
ENST00000380715.1 ENST00000380716.4 ENST00000380738.4 ENST00000380733.4 |
PSIP1 |
PC4 and SFRS1 interacting protein 1 |
| chr9_-_130712995 | 0.16 |
ENST00000373084.4 |
FAM102A |
family with sequence similarity 102, member A |
| chr5_+_138678131 | 0.16 |
ENST00000394795.2 ENST00000510080.1 |
PAIP2 |
poly(A) binding protein interacting protein 2 |
| chr3_+_89156799 | 0.16 |
ENST00000452448.2 ENST00000494014.1 |
EPHA3 |
EPH receptor A3 |
| chr3_+_89156674 | 0.16 |
ENST00000336596.2 |
EPHA3 |
EPH receptor A3 |
| chr2_-_188312971 | 0.16 |
ENST00000410068.1 ENST00000447403.1 ENST00000410102.1 |
CALCRL |
calcitonin receptor-like |
| chr20_-_45984401 | 0.15 |
ENST00000311275.7 |
ZMYND8 |
zinc finger, MYND-type containing 8 |
| chr11_+_111807863 | 0.15 |
ENST00000440460.2 |
DIXDC1 |
DIX domain containing 1 |
| chr6_-_79944336 | 0.15 |
ENST00000344726.5 ENST00000275036.7 |
HMGN3 |
high mobility group nucleosomal binding domain 3 |
| chr1_+_50574585 | 0.14 |
ENST00000371824.1 ENST00000371823.4 |
ELAVL4 |
ELAV like neuron-specific RNA binding protein 4 |
| chr7_+_30960915 | 0.14 |
ENST00000441328.2 ENST00000409899.1 ENST00000409611.1 |
AQP1 |
aquaporin 1 (Colton blood group) |
| chrX_+_107683096 | 0.14 |
ENST00000328300.6 ENST00000361603.2 |
COL4A5 |
collagen, type IV, alpha 5 |
| chr1_-_11751529 | 0.14 |
ENST00000376672.1 |
MAD2L2 |
MAD2 mitotic arrest deficient-like 2 (yeast) |
| chr5_-_38595498 | 0.14 |
ENST00000263409.4 |
LIFR |
leukemia inhibitory factor receptor alpha |
| chr4_+_160188889 | 0.14 |
ENST00000264431.4 |
RAPGEF2 |
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr7_+_28452130 | 0.14 |
ENST00000357727.2 |
CREB5 |
cAMP responsive element binding protein 5 |
| chr7_+_129015484 | 0.14 |
ENST00000490911.1 |
AHCYL2 |
adenosylhomocysteinase-like 2 |
| chr3_+_88188254 | 0.14 |
ENST00000309495.5 |
ZNF654 |
zinc finger protein 654 |
| chr20_+_17207636 | 0.14 |
ENST00000262545.2 |
PCSK2 |
proprotein convertase subtilisin/kexin type 2 |
| chr17_-_33448468 | 0.14 |
ENST00000591723.1 ENST00000593039.1 ENST00000587405.1 |
RAD51L3-RFFL RAD51D |
Uncharacterized protein RAD51 paralog D |
| chr8_-_72274095 | 0.14 |
ENST00000303824.7 |
EYA1 |
eyes absent homolog 1 (Drosophila) |
| chr7_+_134551583 | 0.13 |
ENST00000435928.1 |
CALD1 |
caldesmon 1 |
| chr22_-_36236265 | 0.13 |
ENST00000414461.2 ENST00000416721.2 ENST00000449924.2 ENST00000262829.7 ENST00000397305.3 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr1_-_11751665 | 0.13 |
ENST00000376667.3 ENST00000235310.3 |
MAD2L2 |
MAD2 mitotic arrest deficient-like 2 (yeast) |
| chr4_+_71588372 | 0.13 |
ENST00000536664.1 |
RUFY3 |
RUN and FYVE domain containing 3 |
| chr3_+_141106458 | 0.13 |
ENST00000509883.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr10_+_24528108 | 0.13 |
ENST00000438429.1 |
KIAA1217 |
KIAA1217 |
| chr16_-_31085514 | 0.13 |
ENST00000300849.4 |
ZNF668 |
zinc finger protein 668 |
| chr7_+_111846643 | 0.13 |
ENST00000361822.3 |
ZNF277 |
zinc finger protein 277 |
| chr8_-_57123815 | 0.12 |
ENST00000316981.3 ENST00000423799.2 ENST00000429357.2 |
PLAG1 |
pleiomorphic adenoma gene 1 |
| chr14_-_51027838 | 0.12 |
ENST00000555216.1 |
MAP4K5 |
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr15_-_37391507 | 0.12 |
ENST00000557796.2 ENST00000397620.2 |
MEIS2 |
Meis homeobox 2 |
| chr2_-_2334888 | 0.12 |
ENST00000428368.2 ENST00000399161.2 |
MYT1L |
myelin transcription factor 1-like |
| chr3_-_141747459 | 0.12 |
ENST00000477292.1 ENST00000478006.1 ENST00000495310.1 ENST00000486111.1 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr14_-_69619823 | 0.12 |
ENST00000341516.5 |
DCAF5 |
DDB1 and CUL4 associated factor 5 |
| chrX_-_107682702 | 0.12 |
ENST00000372216.4 |
COL4A6 |
collagen, type IV, alpha 6 |
| chr8_-_70745575 | 0.12 |
ENST00000524945.1 |
SLCO5A1 |
solute carrier organic anion transporter family, member 5A1 |
| chr14_-_55878538 | 0.12 |
ENST00000247178.5 |
ATG14 |
autophagy related 14 |
| chr13_+_103459704 | 0.12 |
ENST00000602836.1 |
BIVM-ERCC5 |
BIVM-ERCC5 readthrough |
| chr10_+_35416223 | 0.12 |
ENST00000489321.1 ENST00000427847.2 ENST00000345491.3 ENST00000395895.2 ENST00000374728.3 ENST00000487132.1 |
CREM |
cAMP responsive element modulator |
| chr19_+_36132631 | 0.11 |
ENST00000379026.2 ENST00000379023.4 ENST00000402764.2 ENST00000479824.1 |
ETV2 |
ets variant 2 |
| chr3_-_141747439 | 0.11 |
ENST00000467667.1 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr9_+_71394945 | 0.11 |
ENST00000394264.3 |
FAM122A |
family with sequence similarity 122A |
| chr14_+_37126765 | 0.11 |
ENST00000402703.2 |
PAX9 |
paired box 9 |
| chr17_+_33448593 | 0.11 |
ENST00000158009.5 |
FNDC8 |
fibronectin type III domain containing 8 |
| chr21_+_35552978 | 0.11 |
ENST00000428914.2 ENST00000609062.1 ENST00000609947.1 |
LINC00310 |
long intergenic non-protein coding RNA 310 |
| chr14_+_23846328 | 0.11 |
ENST00000382809.2 |
CMTM5 |
CKLF-like MARVEL transmembrane domain containing 5 |
| chr4_-_76598544 | 0.11 |
ENST00000515457.1 ENST00000357854.3 |
G3BP2 |
GTPase activating protein (SH3 domain) binding protein 2 |
| chr7_+_134576151 | 0.11 |
ENST00000393118.2 |
CALD1 |
caldesmon 1 |
| chr1_-_193155729 | 0.11 |
ENST00000367434.4 |
B3GALT2 |
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr4_+_113970772 | 0.11 |
ENST00000504454.1 ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2 |
ankyrin 2, neuronal |
| chr19_+_12949251 | 0.10 |
ENST00000251472.4 |
MAST1 |
microtubule associated serine/threonine kinase 1 |
| chr6_+_35996859 | 0.10 |
ENST00000472333.1 |
MAPK14 |
mitogen-activated protein kinase 14 |
| chr3_-_46904946 | 0.10 |
ENST00000292327.4 |
MYL3 |
myosin, light chain 3, alkali; ventricular, skeletal, slow |
| chr11_+_126225529 | 0.10 |
ENST00000227495.6 ENST00000444328.2 ENST00000356132.4 |
ST3GAL4 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr15_-_37391614 | 0.10 |
ENST00000219869.9 |
MEIS2 |
Meis homeobox 2 |
| chr20_+_30555805 | 0.10 |
ENST00000562532.2 |
XKR7 |
XK, Kell blood group complex subunit-related family, member 7 |
| chr3_-_46904918 | 0.10 |
ENST00000395869.1 |
MYL3 |
myosin, light chain 3, alkali; ventricular, skeletal, slow |
| chr10_+_49514698 | 0.10 |
ENST00000432379.1 ENST00000429041.1 ENST00000374189.1 |
MAPK8 |
mitogen-activated protein kinase 8 |
| chr6_+_21666633 | 0.10 |
ENST00000606851.1 |
CASC15 |
cancer susceptibility candidate 15 (non-protein coding) |
| chr1_+_153232160 | 0.10 |
ENST00000368742.3 |
LOR |
loricrin |
| chr5_-_134871639 | 0.10 |
ENST00000314744.4 |
NEUROG1 |
neurogenin 1 |
| chr12_+_32655048 | 0.09 |
ENST00000427716.2 ENST00000266482.3 |
FGD4 |
FYVE, RhoGEF and PH domain containing 4 |
| chr20_+_5987890 | 0.09 |
ENST00000378868.4 |
CRLS1 |
cardiolipin synthase 1 |
| chr7_-_16921601 | 0.09 |
ENST00000402239.3 ENST00000310398.2 ENST00000414935.1 |
AGR3 |
anterior gradient 3 |
| chr9_-_134145880 | 0.09 |
ENST00000372269.3 ENST00000464831.1 |
FAM78A |
family with sequence similarity 78, member A |
| chr7_-_7575477 | 0.09 |
ENST00000399429.3 |
COL28A1 |
collagen, type XXVIII, alpha 1 |
| chr14_+_22977587 | 0.09 |
ENST00000390504.1 |
TRAJ33 |
T cell receptor alpha joining 33 |
| chr1_-_50889155 | 0.08 |
ENST00000404795.3 |
DMRTA2 |
DMRT-like family A2 |
| chr8_+_68864330 | 0.08 |
ENST00000288368.4 |
PREX2 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr1_+_9005917 | 0.08 |
ENST00000549778.1 ENST00000480186.3 ENST00000377443.2 ENST00000377436.3 ENST00000377442.2 |
CA6 |
carbonic anhydrase VI |
| chr3_+_134514093 | 0.08 |
ENST00000398015.3 |
EPHB1 |
EPH receptor B1 |
| chrX_-_112084043 | 0.08 |
ENST00000304758.1 |
AMOT |
angiomotin |
| chr6_-_43027105 | 0.08 |
ENST00000230413.5 ENST00000487429.1 ENST00000489623.1 ENST00000468957.1 |
MRPL2 |
mitochondrial ribosomal protein L2 |
| chr16_+_1832902 | 0.08 |
ENST00000262302.9 ENST00000563136.1 ENST00000565987.1 ENST00000543305.1 ENST00000568287.1 ENST00000565134.1 |
NUBP2 |
nucleotide binding protein 2 |
| chrX_-_13835147 | 0.08 |
ENST00000493677.1 ENST00000355135.2 |
GPM6B |
glycoprotein M6B |
| chr2_-_208989225 | 0.08 |
ENST00000264376.4 |
CRYGD |
crystallin, gamma D |
| chr6_+_43027595 | 0.08 |
ENST00000259708.3 ENST00000472792.1 ENST00000479388.1 ENST00000460283.1 ENST00000394056.2 |
KLC4 |
kinesin light chain 4 |
| chr22_+_43808014 | 0.08 |
ENST00000334209.5 ENST00000443721.1 ENST00000414469.2 ENST00000439548.1 |
MPPED1 |
metallophosphoesterase domain containing 1 |
| chr12_+_60058458 | 0.08 |
ENST00000548610.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr18_+_21594384 | 0.07 |
ENST00000584250.1 |
TTC39C |
tetratricopeptide repeat domain 39C |
| chr7_+_117120017 | 0.07 |
ENST00000003084.6 ENST00000454343.1 |
CFTR |
cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) |
| chr11_+_111808119 | 0.07 |
ENST00000531396.1 |
DIXDC1 |
DIX domain containing 1 |
| chr6_-_133055896 | 0.07 |
ENST00000367927.5 ENST00000425515.2 ENST00000207771.3 ENST00000392393.3 ENST00000450865.2 ENST00000392394.2 |
VNN3 |
vanin 3 |
| chr7_-_135612198 | 0.07 |
ENST00000589735.1 |
LUZP6 |
leucine zipper protein 6 |
| chr14_-_69619291 | 0.07 |
ENST00000554215.1 ENST00000556847.1 |
DCAF5 |
DDB1 and CUL4 associated factor 5 |
| chr1_-_161008697 | 0.07 |
ENST00000318289.10 ENST00000368023.3 ENST00000368024.1 ENST00000423014.2 |
TSTD1 |
thiosulfate sulfurtransferase (rhodanese)-like domain containing 1 |
| chr16_+_2083265 | 0.07 |
ENST00000565855.1 ENST00000566198.1 |
SLC9A3R2 |
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
| chr8_+_57124245 | 0.07 |
ENST00000521831.1 ENST00000355315.3 ENST00000303759.3 ENST00000517636.1 ENST00000517933.1 ENST00000518801.1 ENST00000523975.1 ENST00000396723.5 ENST00000523061.1 ENST00000521524.1 |
CHCHD7 |
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr5_-_146461027 | 0.07 |
ENST00000394410.2 ENST00000508267.1 ENST00000504198.1 |
PPP2R2B |
protein phosphatase 2, regulatory subunit B, beta |
| chr2_-_70475701 | 0.07 |
ENST00000282574.4 |
TIA1 |
TIA1 cytotoxic granule-associated RNA binding protein |
| chr2_+_33661382 | 0.06 |
ENST00000402538.3 |
RASGRP3 |
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr1_-_176176114 | 0.06 |
ENST00000308769.8 |
RFWD2 |
ring finger and WD repeat domain 2, E3 ubiquitin protein ligase |
| chr6_+_50061315 | 0.06 |
ENST00000415106.1 |
RP11-397G17.1 |
RP11-397G17.1 |
| chr1_+_11751748 | 0.06 |
ENST00000294485.5 |
DRAXIN |
dorsal inhibitory axon guidance protein |
| chr4_-_101439148 | 0.06 |
ENST00000511970.1 ENST00000502569.1 ENST00000305864.3 |
EMCN |
endomucin |
| chr12_+_32655110 | 0.06 |
ENST00000546442.1 ENST00000583694.1 |
FGD4 |
FYVE, RhoGEF and PH domain containing 4 |
| chr2_-_70475730 | 0.06 |
ENST00000445587.1 ENST00000433529.2 ENST00000415783.2 |
TIA1 |
TIA1 cytotoxic granule-associated RNA binding protein |
| chr9_-_123476719 | 0.06 |
ENST00000373930.3 |
MEGF9 |
multiple EGF-like-domains 9 |
| chr1_+_43637996 | 0.06 |
ENST00000528956.1 ENST00000529956.1 |
WDR65 |
WD repeat domain 65 |
| chr6_+_43027332 | 0.06 |
ENST00000347162.5 ENST00000453940.2 ENST00000479632.1 ENST00000470728.1 ENST00000458460.2 |
KLC4 |
kinesin light chain 4 |
| chr12_-_110937351 | 0.06 |
ENST00000552130.2 |
VPS29 |
vacuolar protein sorting 29 homolog (S. cerevisiae) |
| chr3_+_25469802 | 0.06 |
ENST00000330688.4 |
RARB |
retinoic acid receptor, beta |
| chr9_-_123476612 | 0.06 |
ENST00000426959.1 |
MEGF9 |
multiple EGF-like-domains 9 |
| chr10_+_52751010 | 0.06 |
ENST00000373985.1 |
PRKG1 |
protein kinase, cGMP-dependent, type I |
| chr1_+_10271674 | 0.06 |
ENST00000377086.1 |
KIF1B |
kinesin family member 1B |
| chr9_-_14314518 | 0.06 |
ENST00000397581.2 |
NFIB |
nuclear factor I/B |
| chr20_-_61002584 | 0.05 |
ENST00000252998.1 |
RBBP8NL |
RBBP8 N-terminal like |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.2 | 1.3 | GO:1903758 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.1 | 0.3 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.1 | 1.0 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 0.3 | GO:0035284 | rhombomere 3 development(GO:0021569) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 0.4 | GO:0030070 | insulin processing(GO:0030070) |
| 0.1 | 0.2 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.1 | 0.5 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 0.3 | GO:0046449 | creatinine metabolic process(GO:0046449) cellular response to UV-A(GO:0071492) |
| 0.1 | 0.2 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.1 | 0.7 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
| 0.1 | 0.2 | GO:0051148 | negative regulation of myoblast differentiation(GO:0045662) negative regulation of muscle cell differentiation(GO:0051148) |
| 0.1 | 0.2 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.1 | 0.5 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:0072229 | carbon dioxide transmembrane transport(GO:0035378) proximal convoluted tubule development(GO:0072019) metanephric proximal convoluted tubule development(GO:0072229) |
| 0.0 | 0.3 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.2 | GO:0030047 | actin modification(GO:0030047) |
| 0.0 | 0.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.4 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.8 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.2 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.0 | 0.1 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.1 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.3 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0035377 | transepithelial water transport(GO:0035377) |
| 0.0 | 0.3 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.3 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.6 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.3 | GO:1900038 | negative regulation of cellular response to hypoxia(GO:1900038) |
| 0.0 | 0.6 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.1 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.2 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0051610 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.1 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.1 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.2 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.4 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.0 | 0.1 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.0 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.0 | 0.1 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.2 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.1 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.9 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 1.0 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 0.2 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 0.1 | GO:1904647 | response to rotenone(GO:1904647) |
| 0.0 | 0.4 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.7 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.3 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.0 | 0.1 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.0 | 0.0 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.1 | GO:0030421 | defecation(GO:0030421) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 5.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.2 | 0.7 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.1 | 0.4 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.1 | GO:0020003 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.2 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.3 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.6 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.1 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 1.3 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.3 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.3 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.8 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.1 | 0.3 | GO:0052839 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.1 | 0.5 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.2 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.0 | 0.1 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.0 | 0.4 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.0 | 0.3 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.8 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.2 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.0 | 1.1 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 1.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.1 | GO:0043337 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.0 | 0.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.1 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.0 | 0.3 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 4.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.6 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.4 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.2 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.0 | 0.6 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.3 | GO:0017091 | AU-rich element binding(GO:0017091) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.5 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 3.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.3 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |