Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
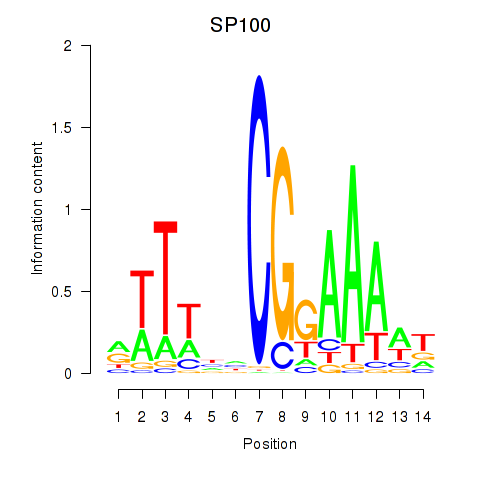
Results for SP100
Z-value: 0.91
Transcription factors associated with SP100
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SP100
|
ENSG00000067066.12 | SP100 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SP100 | hg19_v2_chr2_+_231280954_231280981 | 0.40 | 3.2e-01 | Click! |
Activity profile of SP100 motif
Sorted Z-values of SP100 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SP100
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_155829286 | 0.60 |
ENST00000368324.4 |
SYT11 |
synaptotagmin XI |
| chr14_-_30396948 | 0.48 |
ENST00000331968.5 |
PRKD1 |
protein kinase D1 |
| chr2_+_201450591 | 0.44 |
ENST00000374700.2 |
AOX1 |
aldehyde oxidase 1 |
| chr14_+_29236269 | 0.40 |
ENST00000313071.4 |
FOXG1 |
forkhead box G1 |
| chr7_-_72739575 | 0.39 |
ENST00000453152.1 |
TRIM50 |
tripartite motif containing 50 |
| chr1_+_214776516 | 0.38 |
ENST00000366955.3 |
CENPF |
centromere protein F, 350/400kDa |
| chr10_-_29084886 | 0.37 |
ENST00000608061.1 ENST00000443246.2 ENST00000446012.1 |
LINC00837 |
long intergenic non-protein coding RNA 837 |
| chr11_-_118550375 | 0.34 |
ENST00000525958.1 ENST00000264029.4 ENST00000397925.1 ENST00000529101.1 |
TREH |
trehalase (brush-border membrane glycoprotein) |
| chr17_-_19648916 | 0.32 |
ENST00000444455.1 ENST00000439102.2 |
ALDH3A1 |
aldehyde dehydrogenase 3 family, member A1 |
| chr1_+_90098606 | 0.31 |
ENST00000370454.4 |
LRRC8C |
leucine rich repeat containing 8 family, member C |
| chr13_+_114462193 | 0.30 |
ENST00000375353.3 |
TMEM255B |
transmembrane protein 255B |
| chr16_+_55512742 | 0.29 |
ENST00000568715.1 ENST00000219070.4 |
MMP2 |
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) |
| chr6_+_10585979 | 0.29 |
ENST00000265012.4 |
GCNT2 |
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr10_+_45495898 | 0.29 |
ENST00000298299.3 |
ZNF22 |
zinc finger protein 22 |
| chr4_-_57522598 | 0.28 |
ENST00000553379.2 |
HOPX |
HOP homeobox |
| chr5_+_137514687 | 0.28 |
ENST00000394894.3 |
KIF20A |
kinesin family member 20A |
| chr4_-_57522673 | 0.28 |
ENST00000381255.3 ENST00000317745.7 ENST00000555760.2 ENST00000556614.2 |
HOPX |
HOP homeobox |
| chr2_-_207082748 | 0.28 |
ENST00000407325.2 ENST00000411719.1 |
GPR1 |
G protein-coupled receptor 1 |
| chr4_-_76861392 | 0.27 |
ENST00000505594.1 |
NAAA |
N-acylethanolamine acid amidase |
| chr5_+_72143988 | 0.27 |
ENST00000506351.2 |
TNPO1 |
transportin 1 |
| chr8_+_27629459 | 0.27 |
ENST00000523566.1 |
ESCO2 |
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr10_+_91174314 | 0.26 |
ENST00000371795.4 |
IFIT5 |
interferon-induced protein with tetratricopeptide repeats 5 |
| chr8_+_104383728 | 0.26 |
ENST00000330295.5 |
CTHRC1 |
collagen triple helix repeat containing 1 |
| chrX_+_70586140 | 0.26 |
ENST00000276072.3 |
TAF1 |
TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 250kDa |
| chr2_+_201170703 | 0.25 |
ENST00000358677.5 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
| chr1_-_79472365 | 0.24 |
ENST00000370742.3 |
ELTD1 |
EGF, latrophilin and seven transmembrane domain containing 1 |
| chr11_-_27723158 | 0.23 |
ENST00000395980.2 |
BDNF |
brain-derived neurotrophic factor |
| chr3_-_8811288 | 0.22 |
ENST00000316793.3 ENST00000431493.1 |
OXTR |
oxytocin receptor |
| chr15_+_45879534 | 0.22 |
ENST00000564080.1 ENST00000562384.1 ENST00000569076.1 ENST00000566753.1 |
RP11-96O20.4 BLOC1S6 |
Uncharacterized protein biogenesis of lysosomal organelles complex-1, subunit 6, pallidin |
| chr13_-_114312501 | 0.22 |
ENST00000335288.4 |
ATP4B |
ATPase, H+/K+ exchanging, beta polypeptide |
| chr3_+_111393501 | 0.22 |
ENST00000393934.3 |
PLCXD2 |
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr7_-_91764108 | 0.22 |
ENST00000450723.1 |
CYP51A1 |
cytochrome P450, family 51, subfamily A, polypeptide 1 |
| chrX_-_77395186 | 0.22 |
ENST00000341864.5 |
TAF9B |
TAF9B RNA polymerase II, TATA box binding protein (TBP)-associated factor, 31kDa |
| chr3_-_151047327 | 0.22 |
ENST00000325602.5 |
P2RY13 |
purinergic receptor P2Y, G-protein coupled, 13 |
| chr10_+_60272814 | 0.21 |
ENST00000373886.3 |
BICC1 |
bicaudal C homolog 1 (Drosophila) |
| chr13_+_43597269 | 0.21 |
ENST00000379221.2 |
DNAJC15 |
DnaJ (Hsp40) homolog, subfamily C, member 15 |
| chr8_-_93029865 | 0.20 |
ENST00000422361.2 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr19_-_12267524 | 0.19 |
ENST00000455799.1 ENST00000355738.1 ENST00000439556.2 ENST00000542938.1 |
ZNF625 |
zinc finger protein 625 |
| chr5_+_169010638 | 0.19 |
ENST00000265295.4 ENST00000506574.1 ENST00000515224.1 ENST00000508247.1 ENST00000513941.1 |
SPDL1 |
spindle apparatus coiled-coil protein 1 |
| chr3_-_16524357 | 0.19 |
ENST00000432519.1 |
RFTN1 |
raftlin, lipid raft linker 1 |
| chr14_-_106471723 | 0.18 |
ENST00000390595.2 |
IGHV1-3 |
immunoglobulin heavy variable 1-3 |
| chr5_-_178054014 | 0.18 |
ENST00000520957.1 |
CLK4 |
CDC-like kinase 4 |
| chr5_+_110427983 | 0.18 |
ENST00000513710.2 ENST00000505303.1 |
WDR36 |
WD repeat domain 36 |
| chr5_+_98104978 | 0.18 |
ENST00000308234.7 |
RGMB |
repulsive guidance molecule family member b |
| chr9_-_97402413 | 0.18 |
ENST00000414122.1 |
FBP1 |
fructose-1,6-bisphosphatase 1 |
| chr13_+_111972980 | 0.17 |
ENST00000283547.1 |
TEX29 |
testis expressed 29 |
| chr10_+_89420706 | 0.17 |
ENST00000427144.2 |
PAPSS2 |
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr10_-_4285923 | 0.17 |
ENST00000418372.1 ENST00000608792.1 |
LINC00702 |
long intergenic non-protein coding RNA 702 |
| chr17_-_39538550 | 0.17 |
ENST00000394001.1 |
KRT34 |
keratin 34 |
| chr18_-_24443151 | 0.17 |
ENST00000440832.3 |
AQP4 |
aquaporin 4 |
| chr6_+_151646800 | 0.17 |
ENST00000354675.6 |
AKAP12 |
A kinase (PRKA) anchor protein 12 |
| chrX_-_103268259 | 0.16 |
ENST00000217926.5 |
H2BFWT |
H2B histone family, member W, testis-specific |
| chr16_-_69760409 | 0.16 |
ENST00000561500.1 ENST00000439109.2 ENST00000564043.1 ENST00000379046.2 ENST00000379047.3 |
NQO1 |
NAD(P)H dehydrogenase, quinone 1 |
| chr20_+_5931497 | 0.16 |
ENST00000378886.2 ENST00000265187.4 |
MCM8 |
minichromosome maintenance complex component 8 |
| chr4_-_57976544 | 0.15 |
ENST00000295666.4 ENST00000537922.1 |
IGFBP7 |
insulin-like growth factor binding protein 7 |
| chr17_+_59489112 | 0.15 |
ENST00000335108.2 |
C17orf82 |
chromosome 17 open reading frame 82 |
| chr18_+_3247779 | 0.15 |
ENST00000578611.1 ENST00000583449.1 |
MYL12A |
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr19_+_45147313 | 0.15 |
ENST00000406449.4 |
PVR |
poliovirus receptor |
| chr1_+_109102652 | 0.15 |
ENST00000370035.3 ENST00000405454.1 |
FAM102B |
family with sequence similarity 102, member B |
| chr3_-_108308241 | 0.15 |
ENST00000295746.8 |
KIAA1524 |
KIAA1524 |
| chr17_+_39969183 | 0.15 |
ENST00000321562.4 |
FKBP10 |
FK506 binding protein 10, 65 kDa |
| chr15_-_55881135 | 0.15 |
ENST00000302000.6 |
PYGO1 |
pygopus family PHD finger 1 |
| chr7_+_142985308 | 0.15 |
ENST00000310447.5 |
CASP2 |
caspase 2, apoptosis-related cysteine peptidase |
| chr9_-_97402531 | 0.14 |
ENST00000415431.1 |
FBP1 |
fructose-1,6-bisphosphatase 1 |
| chr3_+_119187785 | 0.14 |
ENST00000295588.4 ENST00000476573.1 |
POGLUT1 |
protein O-glucosyltransferase 1 |
| chr21_-_45078019 | 0.14 |
ENST00000542962.1 |
HSF2BP |
heat shock transcription factor 2 binding protein |
| chr10_+_1095416 | 0.14 |
ENST00000358220.1 |
WDR37 |
WD repeat domain 37 |
| chr15_+_66797627 | 0.14 |
ENST00000565627.1 ENST00000564179.1 |
ZWILCH |
zwilch kinetochore protein |
| chr10_+_37414785 | 0.13 |
ENST00000374660.1 ENST00000602533.1 ENST00000361713.1 |
ANKRD30A |
ankyrin repeat domain 30A |
| chr12_-_10978957 | 0.13 |
ENST00000240619.2 |
TAS2R10 |
taste receptor, type 2, member 10 |
| chr12_+_120884222 | 0.13 |
ENST00000551765.1 ENST00000229384.5 |
GATC |
glutamyl-tRNA(Gln) amidotransferase, subunit C |
| chr11_+_65029233 | 0.13 |
ENST00000265465.3 |
POLA2 |
polymerase (DNA directed), alpha 2, accessory subunit |
| chr4_-_129209944 | 0.13 |
ENST00000520121.1 |
PGRMC2 |
progesterone receptor membrane component 2 |
| chr3_+_122296443 | 0.13 |
ENST00000464300.2 |
PARP15 |
poly (ADP-ribose) polymerase family, member 15 |
| chr2_-_202645612 | 0.13 |
ENST00000409632.2 ENST00000410052.1 ENST00000467448.1 |
ALS2 |
amyotrophic lateral sclerosis 2 (juvenile) |
| chr19_+_44556158 | 0.13 |
ENST00000434772.3 ENST00000585552.1 |
ZNF223 |
zinc finger protein 223 |
| chr8_-_7243080 | 0.13 |
ENST00000400156.4 |
ZNF705G |
zinc finger protein 705G |
| chr15_-_50558223 | 0.13 |
ENST00000267845.3 |
HDC |
histidine decarboxylase |
| chr19_+_56159362 | 0.12 |
ENST00000593069.1 ENST00000308964.3 |
CCDC106 |
coiled-coil domain containing 106 |
| chr20_-_45318230 | 0.12 |
ENST00000372114.3 |
TP53RK |
TP53 regulating kinase |
| chr11_-_5537920 | 0.12 |
ENST00000380184.1 |
UBQLNL |
ubiquilin-like |
| chr2_+_157291953 | 0.12 |
ENST00000310454.6 |
GPD2 |
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr5_+_125935960 | 0.12 |
ENST00000297540.4 |
PHAX |
phosphorylated adaptor for RNA export |
| chr10_-_35379524 | 0.12 |
ENST00000374751.3 ENST00000374742.1 ENST00000602371.1 |
CUL2 |
cullin 2 |
| chr19_+_47852538 | 0.12 |
ENST00000328771.4 |
DHX34 |
DEAH (Asp-Glu-Ala-His) box polypeptide 34 |
| chr3_+_62304712 | 0.12 |
ENST00000494481.1 |
C3orf14 |
chromosome 3 open reading frame 14 |
| chr5_+_54455946 | 0.12 |
ENST00000503787.1 ENST00000296734.6 ENST00000515370.1 |
GPX8 |
glutathione peroxidase 8 (putative) |
| chrX_+_12993202 | 0.12 |
ENST00000451311.2 ENST00000380636.1 |
TMSB4X |
thymosin beta 4, X-linked |
| chr5_+_14143728 | 0.12 |
ENST00000344204.4 ENST00000537187.1 |
TRIO |
trio Rho guanine nucleotide exchange factor |
| chr12_-_58329819 | 0.12 |
ENST00000551421.1 |
RP11-620J15.3 |
RP11-620J15.3 |
| chr2_+_201390843 | 0.12 |
ENST00000357799.4 ENST00000409203.3 |
SGOL2 |
shugoshin-like 2 (S. pombe) |
| chr6_-_27860956 | 0.12 |
ENST00000359611.2 |
HIST1H2AM |
histone cluster 1, H2am |
| chr5_+_151151471 | 0.12 |
ENST00000394123.3 ENST00000543466.1 |
G3BP1 |
GTPase activating protein (SH3 domain) binding protein 1 |
| chr4_-_54232144 | 0.12 |
ENST00000388940.4 ENST00000503450.1 ENST00000401642.3 |
SCFD2 |
sec1 family domain containing 2 |
| chr6_+_96969672 | 0.12 |
ENST00000369278.4 |
UFL1 |
UFM1-specific ligase 1 |
| chrX_+_100353153 | 0.12 |
ENST00000423383.1 ENST00000218507.5 ENST00000403304.2 ENST00000435570.1 |
CENPI |
centromere protein I |
| chr19_-_12512062 | 0.12 |
ENST00000595766.1 ENST00000430385.3 |
ZNF799 |
zinc finger protein 799 |
| chrX_-_57937067 | 0.12 |
ENST00000358697.4 |
ZXDA |
zinc finger, X-linked, duplicated A |
| chr9_-_137809718 | 0.12 |
ENST00000371806.3 |
FCN1 |
ficolin (collagen/fibrinogen domain containing) 1 |
| chr15_+_90777424 | 0.12 |
ENST00000561433.1 ENST00000559204.1 ENST00000558291.1 |
GDPGP1 |
GDP-D-glucose phosphorylase 1 |
| chr1_-_114414316 | 0.11 |
ENST00000528414.1 ENST00000538253.1 ENST00000460620.1 ENST00000420377.2 ENST00000525799.1 ENST00000359785.5 |
PTPN22 |
protein tyrosine phosphatase, non-receptor type 22 (lymphoid) |
| chr12_+_69139886 | 0.11 |
ENST00000398004.2 |
SLC35E3 |
solute carrier family 35, member E3 |
| chr6_+_56911476 | 0.11 |
ENST00000545356.1 |
KIAA1586 |
KIAA1586 |
| chr19_+_58095501 | 0.11 |
ENST00000536878.2 ENST00000597850.1 ENST00000597219.1 ENST00000598689.1 ENST00000599456.1 ENST00000307468.4 |
ZIK1 |
zinc finger protein interacting with K protein 1 |
| chr3_-_156878540 | 0.11 |
ENST00000461804.1 |
CCNL1 |
cyclin L1 |
| chr1_+_227751231 | 0.11 |
ENST00000343776.5 ENST00000608949.1 ENST00000397097.3 |
ZNF678 |
zinc finger protein 678 |
| chr11_-_33744256 | 0.11 |
ENST00000415002.2 ENST00000437761.2 ENST00000445143.2 |
CD59 |
CD59 molecule, complement regulatory protein |
| chr5_+_137514834 | 0.11 |
ENST00000508792.1 ENST00000504621.1 |
KIF20A |
kinesin family member 20A |
| chr7_-_120498357 | 0.11 |
ENST00000415871.1 ENST00000222747.3 ENST00000430985.1 |
TSPAN12 |
tetraspanin 12 |
| chr14_+_105957402 | 0.11 |
ENST00000421892.1 ENST00000334656.7 ENST00000451719.1 ENST00000392522.3 ENST00000392523.4 ENST00000354560.6 ENST00000450383.1 |
C14orf80 |
chromosome 14 open reading frame 80 |
| chr12_+_110011571 | 0.11 |
ENST00000539696.1 ENST00000228510.3 ENST00000392727.3 |
MVK |
mevalonate kinase |
| chr3_-_53916202 | 0.11 |
ENST00000335754.3 |
ACTR8 |
ARP8 actin-related protein 8 homolog (yeast) |
| chr13_-_97646604 | 0.11 |
ENST00000298440.1 ENST00000543457.1 ENST00000541038.1 |
OXGR1 |
oxoglutarate (alpha-ketoglutarate) receptor 1 |
| chr15_+_91446157 | 0.11 |
ENST00000559717.1 |
MAN2A2 |
mannosidase, alpha, class 2A, member 2 |
| chr15_+_66797455 | 0.10 |
ENST00000446801.2 |
ZWILCH |
zwilch kinetochore protein |
| chr5_+_140792614 | 0.10 |
ENST00000398610.2 |
PCDHGA10 |
protocadherin gamma subfamily A, 10 |
| chr1_+_45205498 | 0.10 |
ENST00000372218.4 |
KIF2C |
kinesin family member 2C |
| chr3_-_122134882 | 0.10 |
ENST00000330689.4 |
WDR5B |
WD repeat domain 5B |
| chr11_-_33744487 | 0.10 |
ENST00000426650.2 |
CD59 |
CD59 molecule, complement regulatory protein |
| chr5_+_148651409 | 0.10 |
ENST00000296721.4 |
AFAP1L1 |
actin filament associated protein 1-like 1 |
| chr18_-_32870148 | 0.10 |
ENST00000589178.1 ENST00000333206.5 ENST00000592278.1 ENST00000592211.1 ENST00000420878.3 ENST00000383091.2 ENST00000586922.2 |
ZSCAN30 RP11-158H5.7 |
zinc finger and SCAN domain containing 30 RP11-158H5.7 |
| chr11_-_17410869 | 0.10 |
ENST00000528731.1 |
KCNJ11 |
potassium inwardly-rectifying channel, subfamily J, member 11 |
| chr4_-_52904425 | 0.10 |
ENST00000535450.1 |
SGCB |
sarcoglycan, beta (43kDa dystrophin-associated glycoprotein) |
| chr18_-_43547223 | 0.10 |
ENST00000282041.5 |
EPG5 |
ectopic P-granules autophagy protein 5 homolog (C. elegans) |
| chr20_-_33732952 | 0.10 |
ENST00000541621.1 |
EDEM2 |
ER degradation enhancer, mannosidase alpha-like 2 |
| chr2_+_24346324 | 0.10 |
ENST00000407625.1 ENST00000420135.2 |
FAM228B |
family with sequence similarity 228, member B |
| chr11_+_6947720 | 0.10 |
ENST00000414517.2 |
ZNF215 |
zinc finger protein 215 |
| chr4_+_25378912 | 0.10 |
ENST00000510092.1 ENST00000505991.1 |
ANAPC4 |
anaphase promoting complex subunit 4 |
| chr2_-_39103014 | 0.10 |
ENST00000295373.6 ENST00000417233.1 |
DHX57 |
DEAH (Asp-Glu-Ala-Asp/His) box polypeptide 57 |
| chrX_+_12993336 | 0.10 |
ENST00000380635.1 |
TMSB4X |
thymosin beta 4, X-linked |
| chr2_-_37068530 | 0.10 |
ENST00000593798.1 |
AC007382.1 |
Uncharacterized protein |
| chr6_-_76782371 | 0.10 |
ENST00000369950.3 ENST00000369963.3 |
IMPG1 |
interphotoreceptor matrix proteoglycan 1 |
| chrX_+_11129388 | 0.10 |
ENST00000321143.4 ENST00000380763.3 ENST00000380762.4 |
HCCS |
holocytochrome c synthase |
| chr6_+_57182400 | 0.10 |
ENST00000607273.1 |
PRIM2 |
primase, DNA, polypeptide 2 (58kDa) |
| chr1_-_9131776 | 0.10 |
ENST00000484798.1 |
SLC2A5 |
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr5_+_33440802 | 0.09 |
ENST00000502553.1 ENST00000514259.1 ENST00000265112.3 |
TARS |
threonyl-tRNA synthetase |
| chr4_+_70894130 | 0.09 |
ENST00000526767.1 ENST00000530128.1 ENST00000381057.3 |
HTN3 |
histatin 3 |
| chr16_-_11922665 | 0.09 |
ENST00000573319.1 ENST00000577041.1 ENST00000574028.1 ENST00000571259.1 ENST00000573037.1 ENST00000571158.1 |
BCAR4 |
breast cancer anti-estrogen resistance 4 (non-protein coding) |
| chr6_+_3068606 | 0.09 |
ENST00000259808.4 |
RIPK1 |
receptor (TNFRSF)-interacting serine-threonine kinase 1 |
| chr6_-_41888843 | 0.09 |
ENST00000434077.1 ENST00000409312.1 |
MED20 |
mediator complex subunit 20 |
| chr4_+_25378826 | 0.09 |
ENST00000315368.3 |
ANAPC4 |
anaphase promoting complex subunit 4 |
| chr7_-_156685890 | 0.09 |
ENST00000353442.5 |
LMBR1 |
limb development membrane protein 1 |
| chrX_-_50386648 | 0.09 |
ENST00000460112.3 |
SHROOM4 |
shroom family member 4 |
| chr3_-_150996239 | 0.09 |
ENST00000309170.3 |
P2RY14 |
purinergic receptor P2Y, G-protein coupled, 14 |
| chr2_-_89399845 | 0.09 |
ENST00000479981.1 |
IGKV1-16 |
immunoglobulin kappa variable 1-16 |
| chr8_+_24241969 | 0.09 |
ENST00000522298.1 |
ADAMDEC1 |
ADAM-like, decysin 1 |
| chr6_+_36410762 | 0.09 |
ENST00000483557.1 ENST00000498267.1 ENST00000544295.1 ENST00000449081.2 ENST00000536244.1 ENST00000460983.1 |
KCTD20 |
potassium channel tetramerization domain containing 20 |
| chr10_+_112327425 | 0.09 |
ENST00000361804.4 |
SMC3 |
structural maintenance of chromosomes 3 |
| chr7_+_2443202 | 0.09 |
ENST00000258711.6 |
CHST12 |
carbohydrate (chondroitin 4) sulfotransferase 12 |
| chr2_+_201994569 | 0.09 |
ENST00000457277.1 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chr2_+_158114051 | 0.09 |
ENST00000259056.4 |
GALNT5 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
| chr9_-_33264557 | 0.09 |
ENST00000473781.1 ENST00000488499.1 |
BAG1 |
BCL2-associated athanogene |
| chr4_+_40058411 | 0.09 |
ENST00000261435.6 ENST00000515550.1 |
N4BP2 |
NEDD4 binding protein 2 |
| chr5_+_148651469 | 0.09 |
ENST00000515000.1 |
AFAP1L1 |
actin filament associated protein 1-like 1 |
| chr1_-_156571254 | 0.09 |
ENST00000438976.2 ENST00000334588.7 ENST00000368232.4 ENST00000415314.2 |
GPATCH4 |
G patch domain containing 4 |
| chr11_+_102980126 | 0.09 |
ENST00000375735.2 |
DYNC2H1 |
dynein, cytoplasmic 2, heavy chain 1 |
| chr1_+_225600404 | 0.09 |
ENST00000366845.2 |
AC092811.1 |
AC092811.1 |
| chr19_-_51334769 | 0.09 |
ENST00000596931.1 ENST00000416184.1 ENST00000301421.2 ENST00000598239.1 |
KLK15 |
kallikrein-related peptidase 15 |
| chr11_-_111741994 | 0.09 |
ENST00000398006.2 |
ALG9 |
ALG9, alpha-1,2-mannosyltransferase |
| chr6_-_41888814 | 0.09 |
ENST00000409060.1 ENST00000265350.4 |
MED20 |
mediator complex subunit 20 |
| chr3_+_155838337 | 0.09 |
ENST00000490337.1 ENST00000389636.5 |
KCNAB1 |
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr5_+_151151504 | 0.09 |
ENST00000356245.3 ENST00000507878.2 |
G3BP1 |
GTPase activating protein (SH3 domain) binding protein 1 |
| chr7_-_156685841 | 0.09 |
ENST00000354505.4 ENST00000540390.1 |
LMBR1 |
limb development membrane protein 1 |
| chr11_+_125496400 | 0.09 |
ENST00000524737.1 |
CHEK1 |
checkpoint kinase 1 |
| chr4_+_156588350 | 0.09 |
ENST00000296518.7 |
GUCY1A3 |
guanylate cyclase 1, soluble, alpha 3 |
| chr14_-_39572345 | 0.09 |
ENST00000548032.2 ENST00000556092.1 ENST00000557280.1 ENST00000545328.2 ENST00000553970.1 |
SEC23A |
Sec23 homolog A (S. cerevisiae) |
| chr5_+_140810132 | 0.09 |
ENST00000252085.3 |
PCDHGA12 |
protocadherin gamma subfamily A, 12 |
| chr11_+_125496619 | 0.09 |
ENST00000532669.1 ENST00000278916.3 |
CHEK1 |
checkpoint kinase 1 |
| chr6_-_167571817 | 0.09 |
ENST00000366834.1 |
GPR31 |
G protein-coupled receptor 31 |
| chr10_-_74927810 | 0.08 |
ENST00000372979.4 ENST00000430082.2 ENST00000454759.2 ENST00000413026.1 ENST00000453402.1 |
ECD |
ecdysoneless homolog (Drosophila) |
| chr1_+_13521973 | 0.08 |
ENST00000327795.5 |
PRAMEF21 |
PRAME family member 21 |
| chr3_+_8543393 | 0.08 |
ENST00000157600.3 ENST00000415597.1 ENST00000535732.1 |
LMCD1 |
LIM and cysteine-rich domains 1 |
| chr9_-_135230336 | 0.08 |
ENST00000224140.5 ENST00000372169.2 ENST00000393220.1 |
SETX |
senataxin |
| chr4_+_156588806 | 0.08 |
ENST00000513574.1 |
GUCY1A3 |
guanylate cyclase 1, soluble, alpha 3 |
| chr16_+_16484691 | 0.08 |
ENST00000344087.4 |
NPIPA7 |
nuclear pore complex interacting protein family, member A7 |
| chr6_+_42883727 | 0.08 |
ENST00000304672.1 ENST00000441198.1 ENST00000446507.1 |
PTCRA |
pre T-cell antigen receptor alpha |
| chr2_+_183943464 | 0.08 |
ENST00000354221.4 |
DUSP19 |
dual specificity phosphatase 19 |
| chr10_+_51187938 | 0.08 |
ENST00000311663.5 |
FAM21D |
family with sequence similarity 21, member D |
| chrX_+_36246735 | 0.08 |
ENST00000378653.3 |
CXorf30 |
chromosome X open reading frame 30 |
| chr6_+_109416313 | 0.08 |
ENST00000521277.1 ENST00000517392.1 ENST00000407272.1 ENST00000336977.4 ENST00000519286.1 ENST00000518329.1 ENST00000522461.1 ENST00000518853.1 |
CEP57L1 |
centrosomal protein 57kDa-like 1 |
| chr3_-_197676740 | 0.08 |
ENST00000452735.1 ENST00000453254.1 ENST00000455191.1 |
IQCG |
IQ motif containing G |
| chr19_-_57347415 | 0.08 |
ENST00000601070.1 |
ZIM2 |
zinc finger, imprinted 2 |
| chr4_+_156588249 | 0.08 |
ENST00000393832.3 |
GUCY1A3 |
guanylate cyclase 1, soluble, alpha 3 |
| chr3_+_62304648 | 0.08 |
ENST00000462069.1 ENST00000232519.5 ENST00000465142.1 |
C3orf14 |
chromosome 3 open reading frame 14 |
| chr12_+_95867727 | 0.08 |
ENST00000323666.5 ENST00000546753.1 |
METAP2 |
methionyl aminopeptidase 2 |
| chr4_-_187476721 | 0.08 |
ENST00000307161.5 |
MTNR1A |
melatonin receptor 1A |
| chr10_-_46167722 | 0.08 |
ENST00000374366.3 ENST00000344646.5 |
ZFAND4 |
zinc finger, AN1-type domain 4 |
| chr4_+_184427235 | 0.08 |
ENST00000412117.1 ENST00000434682.2 |
ING2 |
inhibitor of growth family, member 2 |
| chr19_-_53400813 | 0.08 |
ENST00000595635.1 ENST00000594741.1 ENST00000597111.1 ENST00000593618.1 ENST00000597909.1 |
ZNF320 |
zinc finger protein 320 |
| chr1_+_235530675 | 0.08 |
ENST00000366601.3 ENST00000406207.1 ENST00000543662.1 |
TBCE |
tubulin folding cofactor E |
| chr20_+_5107420 | 0.08 |
ENST00000460006.1 |
CDS2 |
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2 |
| chr16_+_50099852 | 0.08 |
ENST00000299192.7 ENST00000285767.4 |
HEATR3 |
HEAT repeat containing 3 |
| chr2_+_201994042 | 0.08 |
ENST00000417748.1 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chr8_+_24241789 | 0.08 |
ENST00000256412.4 ENST00000538205.1 |
ADAMDEC1 |
ADAM-like, decysin 1 |
| chr19_+_37960240 | 0.08 |
ENST00000388801.3 |
ZNF570 |
zinc finger protein 570 |
| chr12_-_91348949 | 0.08 |
ENST00000358859.2 |
CCER1 |
coiled-coil glutamate-rich protein 1 |
| chr8_+_17780483 | 0.08 |
ENST00000517730.1 ENST00000518537.1 ENST00000523055.1 ENST00000519253.1 |
PCM1 |
pericentriolar material 1 |
| chr10_+_92631709 | 0.08 |
ENST00000413330.1 ENST00000277882.3 |
RPP30 |
ribonuclease P/MRP 30kDa subunit |
| chr14_+_50065376 | 0.08 |
ENST00000298288.6 |
LRR1 |
leucine rich repeat protein 1 |
| chr11_+_93474757 | 0.08 |
ENST00000528288.1 |
C11orf54 |
chromosome 11 open reading frame 54 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.2 | GO:1902956 | negative regulation of nucleoside metabolic process(GO:0045978) negative regulation of nucleotide metabolic process(GO:0045980) negative regulation of oxidative phosphorylation(GO:0090324) negative regulation of purine nucleotide metabolic process(GO:1900543) regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902956) negative regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902957) negative regulation of ATP metabolic process(GO:1903579) |
| 0.2 | 0.6 | GO:1990927 | negative regulation of synaptic vesicle recycling(GO:1903422) negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.1 | 0.4 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.1 | 0.3 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.1 | 0.2 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.1 | 0.2 | GO:0034059 | response to anoxia(GO:0034059) |
| 0.1 | 0.5 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.1 | 0.5 | GO:0010767 | regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010767) |
| 0.1 | 0.3 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.2 | GO:0001971 | negative regulation of activation of membrane attack complex(GO:0001971) |
| 0.1 | 0.3 | GO:0052565 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.1 | 0.2 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.1 | 0.2 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.0 | 0.1 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.0 | 0.2 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.1 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.1 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.0 | 0.2 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.1 | GO:0001694 | histamine biosynthetic process(GO:0001694) |
| 0.0 | 0.2 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.1 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.0 | 0.1 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.0 | 0.3 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.3 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.2 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.3 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.1 | GO:0036509 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.0 | 0.3 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.0 | 0.1 | GO:0032242 | regulation of nucleoside transport(GO:0032242) positive regulation of necroptotic process(GO:0060545) |
| 0.0 | 0.1 | GO:0006408 | snRNA export from nucleus(GO:0006408) |
| 0.0 | 0.1 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.1 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 0.2 | GO:0001554 | luteolysis(GO:0001554) |
| 0.0 | 0.4 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.2 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.2 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.0 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.1 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.1 | GO:0031456 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.0 | 0.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.5 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.2 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:0042078 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.2 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.0 | GO:0001978 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.0 | 0.1 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:1990539 | fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.0 | 0.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.1 | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.0 | 0.1 | GO:0051510 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.0 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.1 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.0 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 0.3 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.0 | 0.1 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.0 | 0.3 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.1 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 0.1 | GO:0090212 | vitamin E metabolic process(GO:0042360) regulation of establishment of blood-brain barrier(GO:0090210) negative regulation of establishment of blood-brain barrier(GO:0090212) |
| 0.0 | 0.3 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.0 | 0.0 | GO:0038163 | endomitotic cell cycle(GO:0007113) thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.0 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.0 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.0 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.1 | GO:0036363 | transforming growth factor beta activation(GO:0036363) |
| 0.0 | 0.1 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.0 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.1 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.1 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.1 | GO:1902573 | positive regulation of serine-type endopeptidase activity(GO:1900005) positive regulation of serine-type peptidase activity(GO:1902573) |
| 0.0 | 0.1 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.0 | 0.1 | GO:0032456 | endocytic recycling(GO:0032456) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.2 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.6 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.0 | 0.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.1 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.1 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.1 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.3 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.0 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.0 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.0 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.1 | 0.3 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.1 | 0.2 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 0.3 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.1 | 0.3 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.1 | 0.2 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.1 | 0.2 | GO:0003955 | NAD(P)H dehydrogenase (quinone) activity(GO:0003955) |
| 0.0 | 0.1 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.1 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.3 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.3 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.2 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.2 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.1 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.1 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.1 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.1 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.0 | 0.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.1 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.4 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.1 | GO:0001884 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.5 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.3 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.1 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.2 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.1 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.1 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.0 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.3 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.1 | GO:0086077 | gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.0 | 0.0 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 0.0 | 0.0 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.3 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.6 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.2 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.0 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 0.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.3 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.0 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.0 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.0 | 0.1 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.1 | GO:0019237 | centromeric DNA binding(GO:0019237) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.0 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF MITOTIC PROTEINS | Genes involved in APC/C:Cdc20 mediated degradation of mitotic proteins |
| 0.0 | 0.2 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.3 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.1 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |