Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
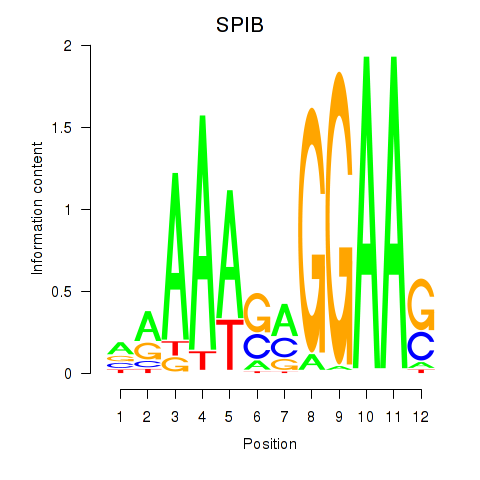
Results for SPIB
Z-value: 0.80
Transcription factors associated with SPIB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SPIB
|
ENSG00000269404.2 | SPIB |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SPIB | hg19_v2_chr19_+_50922187_50922215 | -0.11 | 8.0e-01 | Click! |
Activity profile of SPIB motif
Sorted Z-values of SPIB motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SPIB
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_43886057 | 1.43 |
ENST00000441322.1 ENST00000413657.2 ENST00000453733.1 |
CKMT1B |
creatine kinase, mitochondrial 1B |
| chr15_+_43885252 | 1.35 |
ENST00000453782.1 ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B |
creatine kinase, mitochondrial 1B |
| chr15_+_43985725 | 1.32 |
ENST00000413453.2 |
CKMT1A |
creatine kinase, mitochondrial 1A |
| chr15_+_43985084 | 1.31 |
ENST00000434505.1 ENST00000411750.1 |
CKMT1A |
creatine kinase, mitochondrial 1A |
| chr20_-_46415341 | 1.05 |
ENST00000484875.1 ENST00000361612.4 |
SULF2 |
sulfatase 2 |
| chr20_-_46415297 | 1.03 |
ENST00000467815.1 ENST00000359930.4 |
SULF2 |
sulfatase 2 |
| chr12_-_51785182 | 1.03 |
ENST00000356317.3 ENST00000603188.1 ENST00000604847.1 ENST00000604506.1 |
GALNT6 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 6 (GalNAc-T6) |
| chr17_-_43487780 | 0.90 |
ENST00000532038.1 ENST00000528677.1 |
ARHGAP27 |
Rho GTPase activating protein 27 |
| chr16_+_30194916 | 0.80 |
ENST00000570045.1 ENST00000565497.1 ENST00000570244.1 |
CORO1A |
coronin, actin binding protein, 1A |
| chr11_+_1861399 | 0.79 |
ENST00000381905.3 |
TNNI2 |
troponin I type 2 (skeletal, fast) |
| chrX_+_105937068 | 0.74 |
ENST00000324342.3 |
RNF128 |
ring finger protein 128, E3 ubiquitin protein ligase |
| chr11_+_1860832 | 0.74 |
ENST00000252898.7 |
TNNI2 |
troponin I type 2 (skeletal, fast) |
| chr17_-_56492989 | 0.73 |
ENST00000583753.1 |
RNF43 |
ring finger protein 43 |
| chr9_+_93564039 | 0.72 |
ENST00000375754.4 ENST00000375751.4 |
SYK |
spleen tyrosine kinase |
| chr1_-_175161890 | 0.71 |
ENST00000545251.2 ENST00000423313.1 |
KIAA0040 |
KIAA0040 |
| chr9_+_93564191 | 0.71 |
ENST00000375747.1 |
SYK |
spleen tyrosine kinase |
| chr10_-_98031265 | 0.68 |
ENST00000224337.5 ENST00000371176.2 |
BLNK |
B-cell linker |
| chr9_+_93589734 | 0.66 |
ENST00000375746.1 |
SYK |
spleen tyrosine kinase |
| chr5_+_35856951 | 0.60 |
ENST00000303115.3 ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R |
interleukin 7 receptor |
| chr1_+_205473720 | 0.60 |
ENST00000429964.2 ENST00000506784.1 ENST00000360066.2 |
CDK18 |
cyclin-dependent kinase 18 |
| chr10_-_98031310 | 0.59 |
ENST00000427367.2 ENST00000413476.2 |
BLNK |
B-cell linker |
| chrX_+_138612889 | 0.58 |
ENST00000218099.2 ENST00000394090.2 |
F9 |
coagulation factor IX |
| chr6_-_47010061 | 0.55 |
ENST00000371253.2 |
GPR110 |
G protein-coupled receptor 110 |
| chr2_+_113931513 | 0.52 |
ENST00000245796.6 ENST00000441564.3 |
PSD4 |
pleckstrin and Sec7 domain containing 4 |
| chr15_+_45422131 | 0.52 |
ENST00000321429.4 |
DUOX1 |
dual oxidase 1 |
| chr15_+_45422178 | 0.51 |
ENST00000389037.3 ENST00000558322.1 |
DUOX1 |
dual oxidase 1 |
| chr6_+_116782527 | 0.48 |
ENST00000368606.3 ENST00000368605.1 |
FAM26F |
family with sequence similarity 26, member F |
| chr4_+_103423055 | 0.47 |
ENST00000505458.1 |
NFKB1 |
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| chr5_+_65892174 | 0.46 |
ENST00000404260.3 ENST00000403625.2 ENST00000406374.1 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
| chr19_+_54371114 | 0.45 |
ENST00000448420.1 ENST00000439000.1 ENST00000391770.4 ENST00000391771.1 |
MYADM |
myeloid-associated differentiation marker |
| chr20_+_62492566 | 0.45 |
ENST00000369916.3 |
ABHD16B |
abhydrolase domain containing 16B |
| chr19_-_51471362 | 0.44 |
ENST00000376853.4 ENST00000424910.2 |
KLK6 |
kallikrein-related peptidase 6 |
| chr1_+_161676739 | 0.43 |
ENST00000236938.6 ENST00000367959.2 ENST00000546024.1 ENST00000540521.1 ENST00000367949.2 ENST00000350710.3 ENST00000540926.1 |
FCRLA |
Fc receptor-like A |
| chr19_-_51471381 | 0.42 |
ENST00000594641.1 |
KLK6 |
kallikrein-related peptidase 6 |
| chr17_-_43487741 | 0.41 |
ENST00000455881.1 |
ARHGAP27 |
Rho GTPase activating protein 27 |
| chr1_+_161676983 | 0.41 |
ENST00000367957.2 |
FCRLA |
Fc receptor-like A |
| chrX_-_40594755 | 0.40 |
ENST00000324817.1 |
MED14 |
mediator complex subunit 14 |
| chr1_+_161677034 | 0.39 |
ENST00000349527.4 ENST00000309691.6 ENST00000294796.4 ENST00000367953.3 ENST00000367950.1 |
FCRLA |
Fc receptor-like A |
| chr18_-_52989217 | 0.39 |
ENST00000570287.2 |
TCF4 |
transcription factor 4 |
| chr22_+_37309662 | 0.39 |
ENST00000403662.3 ENST00000262825.5 |
CSF2RB |
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) |
| chr15_+_77287426 | 0.38 |
ENST00000558012.1 ENST00000267939.5 ENST00000379595.3 |
PSTPIP1 |
proline-serine-threonine phosphatase interacting protein 1 |
| chr3_+_98482175 | 0.38 |
ENST00000485391.1 ENST00000492254.1 |
ST3GAL6 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr3_+_121796697 | 0.38 |
ENST00000482356.1 ENST00000393627.2 |
CD86 |
CD86 molecule |
| chr21_-_34186006 | 0.37 |
ENST00000490358.1 |
C21orf62 |
chromosome 21 open reading frame 62 |
| chrX_+_56259316 | 0.37 |
ENST00000468660.1 |
KLF8 |
Kruppel-like factor 8 |
| chr10_-_99161033 | 0.37 |
ENST00000315563.6 ENST00000370992.4 ENST00000414986.1 |
RRP12 |
ribosomal RNA processing 12 homolog (S. cerevisiae) |
| chr2_+_233925064 | 0.36 |
ENST00000359570.5 ENST00000538935.1 |
INPP5D |
inositol polyphosphate-5-phosphatase, 145kDa |
| chr3_-_116163830 | 0.35 |
ENST00000333617.4 |
LSAMP |
limbic system-associated membrane protein |
| chr18_-_52989525 | 0.35 |
ENST00000457482.3 |
TCF4 |
transcription factor 4 |
| chr2_+_113885138 | 0.35 |
ENST00000409930.3 |
IL1RN |
interleukin 1 receptor antagonist |
| chr21_-_34185989 | 0.34 |
ENST00000487113.1 ENST00000382373.4 |
C21orf62 |
chromosome 21 open reading frame 62 |
| chr6_+_130339710 | 0.34 |
ENST00000526087.1 ENST00000533560.1 ENST00000361794.2 |
L3MBTL3 |
l(3)mbt-like 3 (Drosophila) |
| chr19_-_52035044 | 0.34 |
ENST00000359982.4 ENST00000436458.1 ENST00000425629.3 ENST00000391797.3 ENST00000343300.4 |
SIGLEC6 |
sialic acid binding Ig-like lectin 6 |
| chr1_-_120935894 | 0.33 |
ENST00000369383.4 ENST00000369384.4 |
FCGR1B |
Fc fragment of IgG, high affinity Ib, receptor (CD64) |
| chr3_+_46283863 | 0.33 |
ENST00000545097.1 ENST00000541018.1 |
CCR3 |
chemokine (C-C motif) receptor 3 |
| chr3_+_32433363 | 0.33 |
ENST00000465248.1 |
CMTM7 |
CKLF-like MARVEL transmembrane domain containing 7 |
| chr3_+_46283916 | 0.33 |
ENST00000395940.2 |
CCR3 |
chemokine (C-C motif) receptor 3 |
| chr10_-_92617671 | 0.32 |
ENST00000371721.3 |
HTR7 |
5-hydroxytryptamine (serotonin) receptor 7, adenylate cyclase-coupled |
| chr1_+_44440575 | 0.32 |
ENST00000532642.1 ENST00000236067.4 ENST00000471859.2 |
ATP6V0B |
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b |
| chr4_-_87281196 | 0.31 |
ENST00000359221.3 |
MAPK10 |
mitogen-activated protein kinase 10 |
| chr12_+_9102632 | 0.31 |
ENST00000539240.1 |
KLRG1 |
killer cell lectin-like receptor subfamily G, member 1 |
| chr2_+_102972363 | 0.29 |
ENST00000409599.1 |
IL18R1 |
interleukin 18 receptor 1 |
| chr6_-_47009996 | 0.29 |
ENST00000371243.2 |
GPR110 |
G protein-coupled receptor 110 |
| chr8_-_95449155 | 0.28 |
ENST00000481490.2 |
FSBP |
fibrinogen silencer binding protein |
| chr6_+_15246501 | 0.28 |
ENST00000341776.2 |
JARID2 |
jumonji, AT rich interactive domain 2 |
| chr19_+_3136115 | 0.28 |
ENST00000262958.3 |
GNA15 |
guanine nucleotide binding protein (G protein), alpha 15 (Gq class) |
| chr16_+_3096638 | 0.28 |
ENST00000336577.4 |
MMP25 |
matrix metallopeptidase 25 |
| chr1_-_156675368 | 0.28 |
ENST00000368222.3 |
CRABP2 |
cellular retinoic acid binding protein 2 |
| chr12_+_7060432 | 0.27 |
ENST00000318974.9 ENST00000456013.1 |
PTPN6 |
protein tyrosine phosphatase, non-receptor type 6 |
| chr7_+_139529040 | 0.27 |
ENST00000455353.1 ENST00000458722.1 ENST00000411653.1 |
TBXAS1 |
thromboxane A synthase 1 (platelet) |
| chrX_-_152989531 | 0.26 |
ENST00000458587.2 ENST00000416815.1 |
BCAP31 |
B-cell receptor-associated protein 31 |
| chr1_-_160681593 | 0.26 |
ENST00000368045.3 ENST00000368046.3 |
CD48 |
CD48 molecule |
| chr3_-_143567262 | 0.26 |
ENST00000474151.1 ENST00000316549.6 |
SLC9A9 |
solute carrier family 9, subfamily A (NHE9, cation proton antiporter 9), member 9 |
| chr9_-_34397800 | 0.26 |
ENST00000297623.2 |
C9orf24 |
chromosome 9 open reading frame 24 |
| chr3_+_121774202 | 0.26 |
ENST00000469710.1 ENST00000493101.1 ENST00000330540.2 ENST00000264468.5 |
CD86 |
CD86 molecule |
| chr21_-_34185944 | 0.26 |
ENST00000479548.1 |
C21orf62 |
chromosome 21 open reading frame 62 |
| chr17_-_39093672 | 0.25 |
ENST00000209718.3 ENST00000436344.3 ENST00000485751.1 |
KRT23 |
keratin 23 (histone deacetylase inducible) |
| chr8_-_101734170 | 0.25 |
ENST00000522387.1 ENST00000518196.1 |
PABPC1 |
poly(A) binding protein, cytoplasmic 1 |
| chrX_-_152989798 | 0.25 |
ENST00000441714.1 ENST00000442093.1 ENST00000429550.1 ENST00000345046.6 |
BCAP31 |
B-cell receptor-associated protein 31 |
| chr4_-_103266626 | 0.25 |
ENST00000356736.4 |
SLC39A8 |
solute carrier family 39 (zinc transporter), member 8 |
| chr4_+_30721968 | 0.25 |
ENST00000361762.2 |
PCDH7 |
protocadherin 7 |
| chr12_-_15114492 | 0.25 |
ENST00000541546.1 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
| chr1_+_15736359 | 0.24 |
ENST00000375980.4 |
EFHD2 |
EF-hand domain family, member D2 |
| chr7_+_139528952 | 0.24 |
ENST00000416849.2 ENST00000436047.2 ENST00000414508.2 ENST00000448866.1 |
TBXAS1 |
thromboxane A synthase 1 (platelet) |
| chr1_-_23886285 | 0.23 |
ENST00000374561.5 |
ID3 |
inhibitor of DNA binding 3, dominant negative helix-loop-helix protein |
| chr5_-_140013275 | 0.23 |
ENST00000512545.1 ENST00000302014.6 ENST00000401743.2 |
CD14 |
CD14 molecule |
| chr1_+_90287480 | 0.23 |
ENST00000394593.3 |
LRRC8D |
leucine rich repeat containing 8 family, member D |
| chr9_+_112403088 | 0.23 |
ENST00000448454.2 |
PALM2 |
paralemmin 2 |
| chr3_-_20227619 | 0.23 |
ENST00000425061.1 ENST00000443724.1 ENST00000421451.1 ENST00000452020.1 ENST00000417364.1 ENST00000306698.2 ENST00000419233.2 ENST00000263753.4 ENST00000383774.1 ENST00000437051.1 ENST00000412868.1 ENST00000429446.3 ENST00000442720.1 |
SGOL1 |
shugoshin-like 1 (S. pombe) |
| chr7_-_121944491 | 0.23 |
ENST00000331178.4 ENST00000427185.2 ENST00000442488.2 |
FEZF1 |
FEZ family zinc finger 1 |
| chr17_+_7462103 | 0.22 |
ENST00000396545.4 |
TNFSF13 |
tumor necrosis factor (ligand) superfamily, member 13 |
| chr2_-_208031943 | 0.22 |
ENST00000421199.1 ENST00000457962.1 |
KLF7 |
Kruppel-like factor 7 (ubiquitous) |
| chr12_-_8218997 | 0.21 |
ENST00000307637.4 |
C3AR1 |
complement component 3a receptor 1 |
| chr12_-_15114191 | 0.21 |
ENST00000541380.1 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
| chr10_-_62149433 | 0.21 |
ENST00000280772.2 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
| chr3_-_9291063 | 0.21 |
ENST00000383836.3 |
SRGAP3 |
SLIT-ROBO Rho GTPase activating protein 3 |
| chr4_-_80994619 | 0.20 |
ENST00000404191.1 |
ANTXR2 |
anthrax toxin receptor 2 |
| chr2_+_64751433 | 0.20 |
ENST00000238856.4 ENST00000422803.1 ENST00000238855.7 |
AFTPH |
aftiphilin |
| chr13_-_95364389 | 0.20 |
ENST00000376945.2 |
SOX21 |
SRY (sex determining region Y)-box 21 |
| chr6_+_125304502 | 0.19 |
ENST00000519799.1 ENST00000368414.2 ENST00000359704.2 |
RNF217 |
ring finger protein 217 |
| chr9_+_112403059 | 0.19 |
ENST00000374531.2 |
PALM2 |
paralemmin 2 |
| chr1_-_47069886 | 0.19 |
ENST00000371946.4 ENST00000371945.4 ENST00000428112.2 ENST00000529170.1 |
MKNK1 |
MAP kinase interacting serine/threonine kinase 1 |
| chr1_-_43637915 | 0.19 |
ENST00000236051.2 |
EBNA1BP2 |
EBNA1 binding protein 2 |
| chr22_-_37915535 | 0.19 |
ENST00000403299.1 |
CARD10 |
caspase recruitment domain family, member 10 |
| chr1_-_47069955 | 0.19 |
ENST00000341183.5 ENST00000496619.1 |
MKNK1 |
MAP kinase interacting serine/threonine kinase 1 |
| chr2_-_71062938 | 0.19 |
ENST00000410009.3 |
CD207 |
CD207 molecule, langerin |
| chr17_+_7461781 | 0.19 |
ENST00000349228.4 |
TNFSF13 |
tumor necrosis factor (ligand) superfamily, member 13 |
| chr17_+_7461849 | 0.18 |
ENST00000338784.4 |
TNFSF13 |
tumor necrosis factor (ligand) superfamily, member 13 |
| chrX_-_74376108 | 0.18 |
ENST00000339447.4 ENST00000373394.3 ENST00000529949.1 ENST00000534524.1 ENST00000253577.3 |
ABCB7 |
ATP-binding cassette, sub-family B (MDR/TAP), member 7 |
| chr14_+_32546145 | 0.18 |
ENST00000556611.1 ENST00000539826.2 |
ARHGAP5 |
Rho GTPase activating protein 5 |
| chr2_-_242089677 | 0.18 |
ENST00000405260.1 |
PASK |
PAS domain containing serine/threonine kinase |
| chr7_-_36764142 | 0.18 |
ENST00000258749.5 ENST00000535891.1 |
AOAH |
acyloxyacyl hydrolase (neutrophil) |
| chr3_-_116164306 | 0.18 |
ENST00000490035.2 |
LSAMP |
limbic system-associated membrane protein |
| chr21_-_46340884 | 0.17 |
ENST00000302347.5 ENST00000517819.1 |
ITGB2 |
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr10_+_106028605 | 0.17 |
ENST00000450629.2 |
GSTO2 |
glutathione S-transferase omega 2 |
| chr7_+_23636992 | 0.17 |
ENST00000307471.3 ENST00000409765.1 |
CCDC126 |
coiled-coil domain containing 126 |
| chr19_-_3478477 | 0.17 |
ENST00000591708.1 |
C19orf77 |
chromosome 19 open reading frame 77 |
| chr19_-_3479086 | 0.17 |
ENST00000587847.1 |
C19orf77 |
chromosome 19 open reading frame 77 |
| chr8_+_125551338 | 0.17 |
ENST00000276689.3 ENST00000518008.1 ENST00000522532.1 ENST00000517367.1 |
NDUFB9 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 9, 22kDa |
| chr19_+_49838653 | 0.17 |
ENST00000598095.1 ENST00000426897.2 ENST00000323906.4 ENST00000535669.2 ENST00000597602.1 ENST00000595660.1 |
CD37 |
CD37 molecule |
| chr21_-_46340770 | 0.17 |
ENST00000397854.3 |
ITGB2 |
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr19_-_47290535 | 0.17 |
ENST00000412532.2 |
SLC1A5 |
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr4_+_177241094 | 0.17 |
ENST00000503362.1 |
SPCS3 |
signal peptidase complex subunit 3 homolog (S. cerevisiae) |
| chr14_+_75536280 | 0.17 |
ENST00000238686.8 |
ZC2HC1C |
zinc finger, C2HC-type containing 1C |
| chr1_+_112016414 | 0.17 |
ENST00000343534.5 ENST00000369718.3 |
C1orf162 |
chromosome 1 open reading frame 162 |
| chr19_-_3786354 | 0.17 |
ENST00000395040.2 ENST00000310132.6 |
MATK |
megakaryocyte-associated tyrosine kinase |
| chr17_+_65374075 | 0.17 |
ENST00000581322.1 |
PITPNC1 |
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr15_-_73925651 | 0.17 |
ENST00000545878.1 ENST00000287226.8 ENST00000345330.4 |
NPTN |
neuroplastin |
| chr3_-_112693865 | 0.16 |
ENST00000471858.1 ENST00000295863.4 ENST00000308611.3 |
CD200R1 |
CD200 receptor 1 |
| chr11_-_58980342 | 0.16 |
ENST00000361050.3 |
MPEG1 |
macrophage expressed 1 |
| chr2_-_180871780 | 0.16 |
ENST00000410053.3 ENST00000295749.6 ENST00000404136.2 |
CWC22 |
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
| chr1_-_143913143 | 0.16 |
ENST00000400889.1 |
FAM72D |
family with sequence similarity 72, member D |
| chr2_-_113993020 | 0.16 |
ENST00000465084.1 |
PAX8 |
paired box 8 |
| chr14_+_96968707 | 0.16 |
ENST00000216277.8 ENST00000557320.1 ENST00000557471.1 |
PAPOLA |
poly(A) polymerase alpha |
| chr16_+_69373323 | 0.16 |
ENST00000254940.5 |
NIP7 |
NIP7, nucleolar pre-rRNA processing protein |
| chr8_-_101734308 | 0.16 |
ENST00000519004.1 ENST00000519363.1 ENST00000520142.1 |
PABPC1 |
poly(A) binding protein, cytoplasmic 1 |
| chr10_+_106028923 | 0.16 |
ENST00000338595.2 |
GSTO2 |
glutathione S-transferase omega 2 |
| chr1_-_111506562 | 0.16 |
ENST00000485275.2 ENST00000369763.4 |
LRIF1 |
ligand dependent nuclear receptor interacting factor 1 |
| chr16_-_70835034 | 0.16 |
ENST00000261776.5 |
VAC14 |
Vac14 homolog (S. cerevisiae) |
| chr9_+_127624387 | 0.16 |
ENST00000353214.2 |
ARPC5L |
actin related protein 2/3 complex, subunit 5-like |
| chr15_-_33360085 | 0.16 |
ENST00000334528.9 |
FMN1 |
formin 1 |
| chr20_+_30640004 | 0.16 |
ENST00000520553.1 ENST00000518730.1 ENST00000375852.2 |
HCK |
hemopoietic cell kinase |
| chr8_-_74205851 | 0.15 |
ENST00000396467.1 |
RPL7 |
ribosomal protein L7 |
| chr1_+_206138884 | 0.15 |
ENST00000341209.5 ENST00000607379.1 |
FAM72A |
family with sequence similarity 72, member A |
| chr7_-_37488834 | 0.15 |
ENST00000310758.4 |
ELMO1 |
engulfment and cell motility 1 |
| chr9_-_2844058 | 0.15 |
ENST00000397885.2 |
KIAA0020 |
KIAA0020 |
| chr12_-_15114603 | 0.15 |
ENST00000228945.4 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
| chr1_+_206138457 | 0.15 |
ENST00000367128.3 ENST00000431655.2 |
FAM72A |
family with sequence similarity 72, member A |
| chr12_-_55378452 | 0.15 |
ENST00000449076.1 |
TESPA1 |
thymocyte expressed, positive selection associated 1 |
| chr3_+_125694347 | 0.15 |
ENST00000505382.1 ENST00000511082.1 |
ROPN1B |
rhophilin associated tail protein 1B |
| chr8_+_98656693 | 0.15 |
ENST00000519934.1 |
MTDH |
metadherin |
| chr7_+_23637118 | 0.15 |
ENST00000448353.1 |
CCDC126 |
coiled-coil domain containing 126 |
| chr17_-_29641104 | 0.15 |
ENST00000577894.1 ENST00000330927.4 |
EVI2B |
ecotropic viral integration site 2B |
| chr4_-_87281224 | 0.15 |
ENST00000395169.3 ENST00000395161.2 |
MAPK10 |
mitogen-activated protein kinase 10 |
| chrX_+_24072833 | 0.15 |
ENST00000253039.4 |
EIF2S3 |
eukaryotic translation initiation factor 2, subunit 3 gamma, 52kDa |
| chr1_+_120839005 | 0.15 |
ENST00000369390.3 ENST00000452190.1 |
FAM72B |
family with sequence similarity 72, member B |
| chr1_+_120839412 | 0.14 |
ENST00000355228.4 |
FAM72B |
family with sequence similarity 72, member B |
| chr7_+_74188309 | 0.14 |
ENST00000289473.4 ENST00000433458.1 |
NCF1 |
neutrophil cytosolic factor 1 |
| chr1_-_20834586 | 0.14 |
ENST00000264198.3 |
MUL1 |
mitochondrial E3 ubiquitin protein ligase 1 |
| chr1_-_17766198 | 0.14 |
ENST00000375436.4 |
RCC2 |
regulator of chromosome condensation 2 |
| chr14_+_75536335 | 0.14 |
ENST00000554763.1 ENST00000439583.2 ENST00000526130.1 ENST00000525046.1 |
ZC2HC1C |
zinc finger, C2HC-type containing 1C |
| chr13_-_111214015 | 0.14 |
ENST00000267328.3 |
RAB20 |
RAB20, member RAS oncogene family |
| chr12_-_15114658 | 0.14 |
ENST00000542276.1 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
| chr8_-_131028869 | 0.14 |
ENST00000518283.1 ENST00000519110.1 |
FAM49B |
family with sequence similarity 49, member B |
| chr12_+_25205446 | 0.14 |
ENST00000557489.1 ENST00000354454.3 ENST00000536173.1 |
LRMP |
lymphoid-restricted membrane protein |
| chr18_-_32924372 | 0.14 |
ENST00000261332.6 ENST00000399061.3 |
ZNF24 |
zinc finger protein 24 |
| chr14_-_45431091 | 0.14 |
ENST00000579157.1 ENST00000396128.4 ENST00000556500.1 |
KLHL28 |
kelch-like family member 28 |
| chr20_+_62711482 | 0.14 |
ENST00000336866.2 ENST00000355631.4 |
OPRL1 |
opiate receptor-like 1 |
| chr2_-_64751227 | 0.14 |
ENST00000561559.1 |
RP11-568N6.1 |
RP11-568N6.1 |
| chr11_+_55594695 | 0.14 |
ENST00000378397.1 |
OR5L2 |
olfactory receptor, family 5, subfamily L, member 2 |
| chr11_+_46722368 | 0.14 |
ENST00000311764.2 |
ZNF408 |
zinc finger protein 408 |
| chr8_+_125985531 | 0.14 |
ENST00000319286.5 |
ZNF572 |
zinc finger protein 572 |
| chr9_-_95640218 | 0.13 |
ENST00000395506.3 ENST00000375495.3 ENST00000332591.6 |
ZNF484 |
zinc finger protein 484 |
| chr8_-_125551278 | 0.13 |
ENST00000519232.1 ENST00000523888.1 ENST00000522810.1 ENST00000519548.1 ENST00000517678.1 ENST00000605953.1 ENST00000276692.6 |
TATDN1 |
TatD DNase domain containing 1 |
| chr1_+_46713404 | 0.13 |
ENST00000371975.4 ENST00000469835.1 |
RAD54L |
RAD54-like (S. cerevisiae) |
| chr1_+_46713357 | 0.13 |
ENST00000442598.1 |
RAD54L |
RAD54-like (S. cerevisiae) |
| chr7_-_36764004 | 0.13 |
ENST00000431169.1 |
AOAH |
acyloxyacyl hydrolase (neutrophil) |
| chr11_+_60048004 | 0.13 |
ENST00000532114.1 |
MS4A4A |
membrane-spanning 4-domains, subfamily A, member 4A |
| chr16_-_50715196 | 0.13 |
ENST00000423026.2 |
SNX20 |
sorting nexin 20 |
| chr9_-_99145957 | 0.13 |
ENST00000375257.1 ENST00000253270.7 ENST00000375259.4 |
SLC35D2 |
solute carrier family 35 (UDP-GlcNAc/UDP-glucose transporter), member D2 |
| chr10_+_49917751 | 0.13 |
ENST00000325239.5 ENST00000413659.2 |
WDFY4 |
WDFY family member 4 |
| chr2_+_96991935 | 0.13 |
ENST00000361124.4 ENST00000420728.1 ENST00000542887.1 |
ITPRIPL1 |
inositol 1,4,5-trisphosphate receptor interacting protein-like 1 |
| chr11_+_46366918 | 0.13 |
ENST00000528615.1 ENST00000395574.3 |
DGKZ |
diacylglycerol kinase, zeta |
| chr6_+_26251835 | 0.12 |
ENST00000356350.2 |
HIST1H2BH |
histone cluster 1, H2bh |
| chr16_+_30669720 | 0.12 |
ENST00000356166.6 |
FBRS |
fibrosin |
| chr14_-_23288930 | 0.12 |
ENST00000554517.1 ENST00000285850.7 ENST00000397529.2 ENST00000555702.1 |
SLC7A7 |
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr12_+_124086632 | 0.12 |
ENST00000238146.4 ENST00000538744.1 |
DDX55 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 55 |
| chr12_+_54447637 | 0.12 |
ENST00000609810.1 ENST00000430889.2 |
HOXC4 HOXC4 |
homeobox C4 Homeobox protein Hox-C4 |
| chr11_+_60145997 | 0.12 |
ENST00000530614.1 ENST00000530027.1 ENST00000530234.2 ENST00000528215.1 ENST00000531787.1 |
MS4A7 MS4A14 |
membrane-spanning 4-domains, subfamily A, member 7 membrane-spanning 4-domains, subfamily A, member 14 |
| chr3_+_23847394 | 0.12 |
ENST00000306627.3 |
UBE2E1 |
ubiquitin-conjugating enzyme E2E 1 |
| chrX_+_24167828 | 0.12 |
ENST00000379188.3 ENST00000419690.1 ENST00000379177.1 ENST00000304543.5 |
ZFX |
zinc finger protein, X-linked |
| chr5_+_10250328 | 0.12 |
ENST00000515390.1 |
CCT5 |
chaperonin containing TCP1, subunit 5 (epsilon) |
| chr12_+_10365404 | 0.12 |
ENST00000266458.5 ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1 |
GABA(A) receptor-associated protein like 1 |
| chr3_-_112693759 | 0.11 |
ENST00000440122.2 ENST00000490004.1 |
CD200R1 |
CD200 receptor 1 |
| chr22_+_18111594 | 0.11 |
ENST00000399782.1 |
BCL2L13 |
BCL2-like 13 (apoptosis facilitator) |
| chr12_-_122018346 | 0.11 |
ENST00000377069.4 |
KDM2B |
lysine (K)-specific demethylase 2B |
| chr8_-_6420759 | 0.11 |
ENST00000523120.1 |
ANGPT2 |
angiopoietin 2 |
| chr15_-_34629922 | 0.11 |
ENST00000559484.1 ENST00000354181.3 ENST00000558589.1 ENST00000458406.2 |
SLC12A6 |
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr3_-_155462808 | 0.11 |
ENST00000460012.1 |
PLCH1 |
phospholipase C, eta 1 |
| chr6_+_31582961 | 0.11 |
ENST00000376059.3 ENST00000337917.7 |
AIF1 |
allograft inflammatory factor 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:0071226 | response to molecule of fungal origin(GO:0002238) cellular response to molecule of fungal origin(GO:0071226) |
| 0.5 | 2.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.3 | 2.6 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.2 | 0.6 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) |
| 0.2 | 1.0 | GO:0042335 | cuticle development(GO:0042335) |
| 0.1 | 0.9 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.1 | 0.7 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.4 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.1 | 0.4 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.1 | 0.4 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 0.7 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.1 | 0.5 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) |
| 0.1 | 0.3 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.1 | 0.3 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.1 | 0.2 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.1 | 0.7 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.1 | 0.4 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.1 | 0.3 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.1 | 0.2 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.1 | 0.2 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.1 | 0.2 | GO:0071725 | toll-like receptor TLR1:TLR2 signaling pathway(GO:0038123) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.1 | 0.5 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 0.2 | GO:2000612 | thyroid-stimulating hormone secretion(GO:0070460) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.1 | 0.2 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 0.2 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.1 | 0.2 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.1 | 0.5 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.8 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.4 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.5 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.2 | GO:0010585 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.0 | 0.9 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.3 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.2 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.3 | GO:0007207 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.6 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.1 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.1 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.1 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) fourth ventricle development(GO:0021592) |
| 0.0 | 0.3 | GO:0090487 | toxin catabolic process(GO:0009407) secondary metabolite catabolic process(GO:0090487) |
| 0.0 | 0.5 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.5 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.0 | 0.1 | GO:0060454 | positive regulation of gastric acid secretion(GO:0060454) |
| 0.0 | 0.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.2 | GO:0070091 | glucagon secretion(GO:0070091) regulation of glucagon secretion(GO:0070092) |
| 0.0 | 0.1 | GO:0051142 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.0 | 0.3 | GO:0060022 | hard palate development(GO:0060022) |
| 0.0 | 0.3 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.1 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.1 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.5 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0048690 | modulation by virus of host transcription(GO:0019056) regulation of sprouting of injured axon(GO:0048686) positive regulation of sprouting of injured axon(GO:0048687) regulation of axon extension involved in regeneration(GO:0048690) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.0 | 0.1 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.4 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.1 | GO:2000568 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.0 | 0.0 | GO:1900127 | positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.0 | 0.3 | GO:0035655 | interleukin-18-mediated signaling pathway(GO:0035655) |
| 0.0 | 0.2 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.1 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) |
| 0.0 | 0.1 | GO:0000354 | cis assembly of pre-catalytic spliceosome(GO:0000354) |
| 0.0 | 0.1 | GO:0042040 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.0 | 0.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.1 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.0 | 0.1 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 0.0 | 0.3 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.0 | 0.4 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 1.2 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.3 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.2 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.1 | GO:0048704 | embryonic cranial skeleton morphogenesis(GO:0048701) embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 0.3 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.2 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.2 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.1 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.0 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.1 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.1 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.0 | 0.1 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 1.0 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.2 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.2 | GO:0030903 | notochord development(GO:0030903) |
| 0.0 | 0.1 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.1 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.9 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.3 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.2 | GO:0006465 | signal peptide processing(GO:0006465) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.8 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 0.5 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.1 | 1.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.4 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.1 | 0.4 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 0.2 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.5 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.2 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.3 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.9 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.3 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.2 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.1 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.1 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.0 | 2.1 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.2 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.0 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.6 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.3 | 2.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 0.5 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.1 | 0.4 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.1 | 2.1 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.1 | 0.3 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.1 | 1.5 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.1 | 0.3 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.1 | 0.4 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.1 | 0.5 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 1.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 0.4 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.1 | 0.7 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 1.3 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.4 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 0.7 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.3 | GO:0050610 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.1 | 0.7 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.4 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.0 | 0.8 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.2 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.0 | 0.2 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 1.0 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.0 | 0.4 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.3 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.2 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.2 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.2 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.0 | 0.1 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.0 | 0.4 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.3 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.3 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.0 | 0.4 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.2 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.5 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.1 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.0 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.7 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.3 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.0 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.4 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 1.1 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.9 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.1 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.8 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.7 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.6 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 0.5 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 0.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.6 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.6 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.2 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.3 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.5 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 1.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.4 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 5.4 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 1.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |