Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
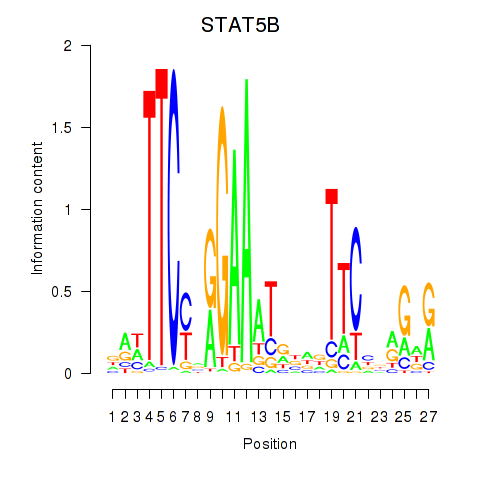
Results for STAT5B
Z-value: 1.32
Transcription factors associated with STAT5B
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
STAT5B
|
ENSG00000173757.5 | STAT5B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| STAT5B | hg19_v2_chr17_-_40428359_40428462 | -0.66 | 7.8e-02 | Click! |
Activity profile of STAT5B motif
Sorted Z-values of STAT5B motif
Network of associatons between targets according to the STRING database.
First level regulatory network of STAT5B
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_154250989 | 1.70 |
ENST00000360256.4 |
F8 |
coagulation factor VIII, procoagulant component |
| chr5_+_178450753 | 1.58 |
ENST00000444149.2 ENST00000519896.1 ENST00000522442.1 |
ZNF879 |
zinc finger protein 879 |
| chr5_-_111093081 | 1.56 |
ENST00000453526.2 ENST00000509427.1 |
NREP |
neuronal regeneration related protein |
| chr5_-_111092873 | 1.47 |
ENST00000509025.1 ENST00000515855.1 |
NREP |
neuronal regeneration related protein |
| chr19_+_35168633 | 1.43 |
ENST00000505365.2 |
ZNF302 |
zinc finger protein 302 |
| chr19_+_35168567 | 1.37 |
ENST00000457781.2 ENST00000505163.1 ENST00000505242.1 ENST00000423823.2 ENST00000507959.1 ENST00000446502.2 |
ZNF302 |
zinc finger protein 302 |
| chr19_+_35168547 | 1.36 |
ENST00000502743.1 ENST00000509528.1 ENST00000506901.1 |
ZNF302 |
zinc finger protein 302 |
| chr19_-_44860820 | 1.34 |
ENST00000354340.4 ENST00000337401.4 ENST00000587909.1 |
ZNF112 |
zinc finger protein 112 |
| chr5_-_111092930 | 1.30 |
ENST00000257435.7 |
NREP |
neuronal regeneration related protein |
| chr5_-_111093340 | 1.27 |
ENST00000508870.1 |
NREP |
neuronal regeneration related protein |
| chr5_-_111093167 | 1.24 |
ENST00000446294.2 ENST00000419114.2 |
NREP |
neuronal regeneration related protein |
| chr19_-_44809121 | 1.12 |
ENST00000591609.1 ENST00000589799.1 ENST00000291182.4 ENST00000589248.1 |
ZNF235 |
zinc finger protein 235 |
| chr19_+_44669280 | 1.10 |
ENST00000590089.1 ENST00000454662.2 |
ZNF226 |
zinc finger protein 226 |
| chr19_-_44405941 | 0.91 |
ENST00000587128.1 |
RP11-15A1.3 |
RP11-15A1.3 |
| chr19_-_44405623 | 0.78 |
ENST00000591815.1 |
RP11-15A1.3 |
RP11-15A1.3 |
| chr19_-_37958318 | 0.71 |
ENST00000316950.6 ENST00000591710.1 |
ZNF569 |
zinc finger protein 569 |
| chr19_-_58090240 | 0.69 |
ENST00000196489.3 |
ZNF416 |
zinc finger protein 416 |
| chr19_+_44617511 | 0.69 |
ENST00000262894.6 ENST00000588926.1 ENST00000592780.1 |
ZNF225 |
zinc finger protein 225 |
| chr19_+_44764031 | 0.68 |
ENST00000592581.1 ENST00000590668.1 ENST00000588489.1 ENST00000391958.2 |
ZNF233 |
zinc finger protein 233 |
| chr5_-_95158375 | 0.68 |
ENST00000512469.2 ENST00000379979.4 ENST00000505427.1 ENST00000508780.1 |
GLRX |
glutaredoxin (thioltransferase) |
| chr19_-_37958086 | 0.68 |
ENST00000592490.1 ENST00000392149.2 |
ZNF569 |
zinc finger protein 569 |
| chr5_-_95158644 | 0.68 |
ENST00000237858.6 |
GLRX |
glutaredoxin (thioltransferase) |
| chr19_+_44598534 | 0.68 |
ENST00000336976.6 |
ZNF224 |
zinc finger protein 224 |
| chr19_+_44529479 | 0.67 |
ENST00000587846.1 ENST00000187879.8 ENST00000391960.3 |
ZNF222 |
zinc finger protein 222 |
| chr19_+_44507091 | 0.65 |
ENST00000429154.2 ENST00000585632.1 |
ZNF230 |
zinc finger protein 230 |
| chr19_+_35225060 | 0.64 |
ENST00000599244.1 ENST00000392232.3 |
ZNF181 |
zinc finger protein 181 |
| chr16_-_89785777 | 0.63 |
ENST00000561976.1 |
VPS9D1 |
VPS9 domain containing 1 |
| chr19_+_44556158 | 0.62 |
ENST00000434772.3 ENST00000585552.1 |
ZNF223 |
zinc finger protein 223 |
| chr3_-_183735651 | 0.62 |
ENST00000427120.2 ENST00000392579.2 ENST00000382494.2 ENST00000446941.2 |
ABCC5 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr3_+_130745769 | 0.58 |
ENST00000412440.2 |
NEK11 |
NIMA-related kinase 11 |
| chr3_+_180319918 | 0.57 |
ENST00000296015.4 ENST00000491380.1 ENST00000412756.2 ENST00000382584.4 |
TTC14 |
tetratricopeptide repeat domain 14 |
| chr14_-_73493784 | 0.57 |
ENST00000553891.1 |
ZFYVE1 |
zinc finger, FYVE domain containing 1 |
| chr2_-_202563414 | 0.56 |
ENST00000409474.3 ENST00000315506.7 ENST00000359962.5 |
MPP4 |
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4) |
| chr19_-_44439387 | 0.54 |
ENST00000269973.5 |
ZNF45 |
zinc finger protein 45 |
| chr2_-_202562716 | 0.53 |
ENST00000428900.2 |
MPP4 |
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4) |
| chr19_+_44645700 | 0.53 |
ENST00000592437.1 |
ZNF234 |
zinc finger protein 234 |
| chr14_-_73493825 | 0.53 |
ENST00000318876.5 ENST00000556143.1 |
ZFYVE1 |
zinc finger, FYVE domain containing 1 |
| chr15_+_91643442 | 0.52 |
ENST00000394232.1 |
SV2B |
synaptic vesicle glycoprotein 2B |
| chr5_-_177423243 | 0.51 |
ENST00000308304.2 |
PROP1 |
PROP paired-like homeobox 1 |
| chr1_-_109618566 | 0.51 |
ENST00000338366.5 |
TAF13 |
TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa |
| chr6_-_27440837 | 0.51 |
ENST00000211936.6 |
ZNF184 |
zinc finger protein 184 |
| chr6_-_27440460 | 0.50 |
ENST00000377419.1 |
ZNF184 |
zinc finger protein 184 |
| chr9_+_104161123 | 0.50 |
ENST00000374861.3 ENST00000339664.2 ENST00000259395.4 |
ZNF189 |
zinc finger protein 189 |
| chr19_+_37709076 | 0.49 |
ENST00000590503.1 ENST00000589413.1 |
ZNF383 |
zinc finger protein 383 |
| chr8_+_38586068 | 0.48 |
ENST00000443286.2 ENST00000520340.1 ENST00000518415.1 |
TACC1 |
transforming, acidic coiled-coil containing protein 1 |
| chr12_+_133657461 | 0.47 |
ENST00000412146.2 ENST00000544426.1 ENST00000440984.2 ENST00000319849.3 ENST00000440550.2 |
ZNF140 |
zinc finger protein 140 |
| chr19_+_38085731 | 0.47 |
ENST00000589117.1 |
ZNF540 |
zinc finger protein 540 |
| chr17_-_18585131 | 0.46 |
ENST00000443457.1 ENST00000583002.1 |
ZNF286B |
zinc finger protein 286B |
| chr3_+_130745688 | 0.43 |
ENST00000510769.1 ENST00000429253.2 ENST00000356918.4 ENST00000510688.1 ENST00000511262.1 ENST00000383366.4 |
NEK11 |
NIMA-related kinase 11 |
| chr5_-_178157700 | 0.42 |
ENST00000335815.2 |
ZNF354A |
zinc finger protein 354A |
| chr19_+_44488330 | 0.42 |
ENST00000591532.1 ENST00000407951.2 ENST00000270014.2 ENST00000590615.1 ENST00000586454.1 |
ZNF155 |
zinc finger protein 155 |
| chr19_+_58258164 | 0.42 |
ENST00000317178.5 |
ZNF776 |
zinc finger protein 776 |
| chr17_-_16472483 | 0.42 |
ENST00000395824.1 ENST00000448349.2 ENST00000395825.3 |
ZNF287 |
zinc finger protein 287 |
| chrX_+_106045891 | 0.42 |
ENST00000357242.5 ENST00000310452.2 ENST00000481617.2 ENST00000276175.3 |
TBC1D8B |
TBC1 domain family, member 8B (with GRAM domain) |
| chr7_+_99102267 | 0.41 |
ENST00000326775.5 ENST00000451158.1 |
ZKSCAN5 |
zinc finger with KRAB and SCAN domains 5 |
| chr5_+_178286925 | 0.41 |
ENST00000322434.3 |
ZNF354B |
zinc finger protein 354B |
| chr19_+_37407212 | 0.41 |
ENST00000427117.1 ENST00000587130.1 ENST00000333987.7 ENST00000415168.1 ENST00000444991.1 |
ZNF568 |
zinc finger protein 568 |
| chr12_-_133532864 | 0.40 |
ENST00000536932.1 ENST00000360187.4 ENST00000392321.3 |
CHFR ZNF605 |
checkpoint with forkhead and ring finger domains, E3 ubiquitin protein ligase zinc finger protein 605 |
| chr17_-_80291818 | 0.38 |
ENST00000269389.3 ENST00000581691.1 |
SECTM1 |
secreted and transmembrane 1 |
| chr11_-_55703876 | 0.38 |
ENST00000301532.3 |
OR5I1 |
olfactory receptor, family 5, subfamily I, member 1 |
| chr7_+_142829162 | 0.37 |
ENST00000291009.3 |
PIP |
prolactin-induced protein |
| chr7_+_99102573 | 0.37 |
ENST00000394170.2 |
ZKSCAN5 |
zinc finger with KRAB and SCAN domains 5 |
| chr19_-_36980455 | 0.37 |
ENST00000454319.1 ENST00000392170.2 |
ZNF566 |
zinc finger protein 566 |
| chr13_-_99404875 | 0.37 |
ENST00000376503.5 |
SLC15A1 |
solute carrier family 15 (oligopeptide transporter), member 1 |
| chr19_-_36980337 | 0.36 |
ENST00000434377.2 ENST00000424129.2 ENST00000452939.1 ENST00000427002.1 |
ZNF566 |
zinc finger protein 566 |
| chr11_-_82997013 | 0.36 |
ENST00000529073.1 ENST00000529611.1 |
CCDC90B |
coiled-coil domain containing 90B |
| chr19_-_40023450 | 0.36 |
ENST00000326282.4 |
EID2B |
EP300 interacting inhibitor of differentiation 2B |
| chr19_+_44716678 | 0.35 |
ENST00000586228.1 ENST00000588219.1 ENST00000313040.7 ENST00000589707.1 ENST00000588394.1 ENST00000589005.1 |
ZNF227 |
zinc finger protein 227 |
| chr11_-_82997420 | 0.34 |
ENST00000455220.2 ENST00000529689.1 |
CCDC90B |
coiled-coil domain containing 90B |
| chr11_+_2920951 | 0.34 |
ENST00000347936.2 |
SLC22A18 |
solute carrier family 22, member 18 |
| chr19_+_44669221 | 0.34 |
ENST00000590578.1 ENST00000589160.1 ENST00000337433.5 ENST00000586286.1 ENST00000585560.1 ENST00000586914.1 ENST00000588883.1 ENST00000413984.2 ENST00000588742.1 ENST00000300823.6 ENST00000585678.1 ENST00000586203.1 ENST00000590467.1 ENST00000588795.1 ENST00000588127.1 |
ZNF226 |
zinc finger protein 226 |
| chr19_-_58220517 | 0.34 |
ENST00000512439.2 ENST00000426889.1 |
ZNF154 |
zinc finger protein 154 |
| chr19_-_45004556 | 0.34 |
ENST00000587047.1 ENST00000391956.4 ENST00000221327.4 ENST00000586637.1 ENST00000591064.1 ENST00000592529.1 |
ZNF180 |
zinc finger protein 180 |
| chr19_-_38085633 | 0.34 |
ENST00000593133.1 ENST00000590751.1 ENST00000358744.3 ENST00000328550.2 ENST00000451802.2 |
ZNF571 |
zinc finger protein 571 |
| chr1_+_202431859 | 0.33 |
ENST00000391959.3 ENST00000367270.4 |
PPP1R12B |
protein phosphatase 1, regulatory subunit 12B |
| chr2_-_27886676 | 0.33 |
ENST00000337768.5 |
SUPT7L |
suppressor of Ty 7 (S. cerevisiae)-like |
| chr6_+_29429217 | 0.33 |
ENST00000396792.2 |
OR2H1 |
olfactory receptor, family 2, subfamily H, member 1 |
| chr16_-_18462221 | 0.32 |
ENST00000528301.1 |
RP11-1212A22.4 |
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr19_+_38085768 | 0.32 |
ENST00000316433.4 ENST00000590588.1 ENST00000586134.1 ENST00000586792.1 |
ZNF540 |
zinc finger protein 540 |
| chr19_-_50529193 | 0.32 |
ENST00000596445.1 ENST00000599538.1 |
VRK3 |
vaccinia related kinase 3 |
| chr19_+_10197463 | 0.31 |
ENST00000590378.1 ENST00000397881.3 |
C19orf66 |
chromosome 19 open reading frame 66 |
| chr19_-_36980756 | 0.31 |
ENST00000493391.1 |
ZNF566 |
zinc finger protein 566 |
| chr11_-_82997371 | 0.31 |
ENST00000525503.1 |
CCDC90B |
coiled-coil domain containing 90B |
| chr9_+_130186653 | 0.31 |
ENST00000342483.5 ENST00000543471.1 |
ZNF79 |
zinc finger protein 79 |
| chr8_-_146012660 | 0.31 |
ENST00000343459.4 ENST00000429371.2 ENST00000534445.1 |
ZNF34 |
zinc finger protein 34 |
| chr15_+_90895471 | 0.31 |
ENST00000354377.3 ENST00000379090.5 |
ZNF774 |
zinc finger protein 774 |
| chr16_+_16434185 | 0.30 |
ENST00000524823.2 |
AC138969.4 |
Protein PKD1P1 |
| chr2_-_63815628 | 0.30 |
ENST00000409562.3 |
WDPCP |
WD repeat containing planar cell polarity effector |
| chr15_+_67420441 | 0.30 |
ENST00000558894.1 |
SMAD3 |
SMAD family member 3 |
| chr19_-_37406923 | 0.30 |
ENST00000520965.1 |
ZNF829 |
zinc finger protein 829 |
| chr8_+_17780483 | 0.29 |
ENST00000517730.1 ENST00000518537.1 ENST00000523055.1 ENST00000519253.1 |
PCM1 |
pericentriolar material 1 |
| chr1_+_153631130 | 0.29 |
ENST00000368685.5 |
SNAPIN |
SNAP-associated protein |
| chr7_+_142985308 | 0.29 |
ENST00000310447.5 |
CASP2 |
caspase 2, apoptosis-related cysteine peptidase |
| chr9_+_112542591 | 0.28 |
ENST00000483909.1 ENST00000314527.4 ENST00000413420.1 ENST00000302798.7 ENST00000555236.1 ENST00000510514.5 |
PALM2 PALM2-AKAP2 AKAP2 |
paralemmin 2 PALM2-AKAP2 readthrough A kinase (PRKA) anchor protein 2 |
| chr9_+_100069933 | 0.28 |
ENST00000529487.1 |
CCDC180 |
coiled-coil domain containing 180 |
| chr3_-_151047327 | 0.28 |
ENST00000325602.5 |
P2RY13 |
purinergic receptor P2Y, G-protein coupled, 13 |
| chr9_-_115774453 | 0.28 |
ENST00000427548.1 |
ZNF883 |
zinc finger protein 883 |
| chr5_-_137514333 | 0.28 |
ENST00000411594.2 ENST00000430331.1 |
BRD8 |
bromodomain containing 8 |
| chr11_+_71640071 | 0.28 |
ENST00000533380.1 ENST00000393713.3 ENST00000545854.1 |
RNF121 |
ring finger protein 121 |
| chr14_-_20881579 | 0.27 |
ENST00000556935.1 ENST00000262715.5 ENST00000556549.1 |
TEP1 |
telomerase-associated protein 1 |
| chr3_-_64211112 | 0.27 |
ENST00000295902.6 |
PRICKLE2 |
prickle homolog 2 (Drosophila) |
| chr19_+_44645731 | 0.27 |
ENST00000426739.2 |
ZNF234 |
zinc finger protein 234 |
| chr10_-_127505167 | 0.26 |
ENST00000368786.1 |
UROS |
uroporphyrinogen III synthase |
| chr7_-_151107106 | 0.26 |
ENST00000334493.6 |
WDR86 |
WD repeat domain 86 |
| chr19_+_40502938 | 0.26 |
ENST00000599504.1 ENST00000596894.1 ENST00000601138.1 ENST00000600094.1 ENST00000347077.4 |
ZNF546 |
zinc finger protein 546 |
| chr11_+_71640112 | 0.26 |
ENST00000530137.1 |
RNF121 |
ring finger protein 121 |
| chr5_-_137514288 | 0.26 |
ENST00000454473.1 ENST00000418329.1 ENST00000455658.2 ENST00000230901.5 ENST00000402931.1 |
BRD8 |
bromodomain containing 8 |
| chr22_-_37213554 | 0.26 |
ENST00000443735.1 |
PVALB |
parvalbumin |
| chr12_-_46384334 | 0.24 |
ENST00000369367.3 ENST00000266589.6 ENST00000395453.2 ENST00000395454.2 |
SCAF11 |
SR-related CTD-associated factor 11 |
| chr4_+_74347400 | 0.24 |
ENST00000226355.3 |
AFM |
afamin |
| chr1_+_93646238 | 0.24 |
ENST00000448243.1 ENST00000370276.1 |
CCDC18 |
coiled-coil domain containing 18 |
| chr2_-_63815860 | 0.24 |
ENST00000272321.7 ENST00000431065.1 |
WDPCP |
WD repeat containing planar cell polarity effector |
| chr14_-_70883708 | 0.23 |
ENST00000256366.4 |
SYNJ2BP |
synaptojanin 2 binding protein |
| chr2_-_231084617 | 0.23 |
ENST00000409815.2 |
SP110 |
SP110 nuclear body protein |
| chr7_-_151107767 | 0.23 |
ENST00000477459.1 |
WDR86 |
WD repeat domain 86 |
| chr1_-_146054494 | 0.22 |
ENST00000401009.2 |
NBPF11 |
neuroblastoma breakpoint family, member 11 |
| chr7_-_6098770 | 0.22 |
ENST00000536084.1 ENST00000446699.1 ENST00000199389.6 |
EIF2AK1 |
eukaryotic translation initiation factor 2-alpha kinase 1 |
| chr19_+_37341260 | 0.22 |
ENST00000589046.1 |
ZNF345 |
zinc finger protein 345 |
| chr4_+_25378912 | 0.21 |
ENST00000510092.1 ENST00000505991.1 |
ANAPC4 |
anaphase promoting complex subunit 4 |
| chr19_+_44576296 | 0.21 |
ENST00000421176.3 |
ZNF284 |
zinc finger protein 284 |
| chr17_+_5323492 | 0.21 |
ENST00000405578.4 ENST00000574003.1 |
RPAIN |
RPA interacting protein |
| chr7_-_93633658 | 0.21 |
ENST00000433727.1 |
BET1 |
Bet1 golgi vesicular membrane trafficking protein |
| chr4_+_25378826 | 0.21 |
ENST00000315368.3 |
ANAPC4 |
anaphase promoting complex subunit 4 |
| chr7_-_93633684 | 0.21 |
ENST00000222547.3 ENST00000425626.1 |
BET1 |
Bet1 golgi vesicular membrane trafficking protein |
| chr20_+_39657454 | 0.21 |
ENST00000361337.2 |
TOP1 |
topoisomerase (DNA) I |
| chr9_-_70488865 | 0.20 |
ENST00000377392.5 |
CBWD5 |
COBW domain containing 5 |
| chr2_-_27886460 | 0.20 |
ENST00000404798.2 ENST00000405491.1 ENST00000464789.2 ENST00000406540.1 |
SUPT7L |
suppressor of Ty 7 (S. cerevisiae)-like |
| chr19_+_45445491 | 0.20 |
ENST00000592954.1 ENST00000419266.2 ENST00000589057.1 |
APOC4 APOC4-APOC2 |
apolipoprotein C-IV APOC4-APOC2 readthrough (NMD candidate) |
| chr16_-_70472946 | 0.20 |
ENST00000342907.2 |
ST3GAL2 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 2 |
| chr3_-_150920979 | 0.19 |
ENST00000309180.5 ENST00000480322.1 |
GPR171 |
G protein-coupled receptor 171 |
| chr19_+_37407351 | 0.19 |
ENST00000455427.2 ENST00000587857.1 |
ZNF568 |
zinc finger protein 568 |
| chr11_-_62911693 | 0.19 |
ENST00000417740.1 ENST00000326192.5 |
SLC22A24 |
solute carrier family 22, member 24 |
| chr13_+_115079949 | 0.19 |
ENST00000361283.1 |
CHAMP1 |
chromosome alignment maintaining phosphoprotein 1 |
| chr19_+_58361185 | 0.19 |
ENST00000339656.5 ENST00000423137.1 |
ZNF587 |
zinc finger protein 587 |
| chr5_+_137514834 | 0.19 |
ENST00000508792.1 ENST00000504621.1 |
KIF20A |
kinesin family member 20A |
| chr19_-_57988871 | 0.19 |
ENST00000596831.1 ENST00000601768.1 ENST00000356584.3 ENST00000600175.1 ENST00000425074.3 ENST00000343280.4 ENST00000427512.2 |
AC004076.9 ZNF772 |
Uncharacterized protein zinc finger protein 772 |
| chr19_+_39881951 | 0.18 |
ENST00000315588.5 ENST00000594368.1 ENST00000599213.2 ENST00000596297.1 |
MED29 |
mediator complex subunit 29 |
| chr4_+_86396265 | 0.18 |
ENST00000395184.1 |
ARHGAP24 |
Rho GTPase activating protein 24 |
| chr7_-_26904317 | 0.18 |
ENST00000345317.2 |
SKAP2 |
src kinase associated phosphoprotein 2 |
| chr1_-_54519134 | 0.18 |
ENST00000371341.1 |
TMEM59 |
transmembrane protein 59 |
| chr6_+_52226897 | 0.18 |
ENST00000442253.2 |
PAQR8 |
progestin and adipoQ receptor family member VIII |
| chr3_-_119379719 | 0.18 |
ENST00000493094.1 |
POPDC2 |
popeye domain containing 2 |
| chr5_+_137514687 | 0.17 |
ENST00000394894.3 |
KIF20A |
kinesin family member 20A |
| chr19_-_37407172 | 0.17 |
ENST00000391711.3 |
ZNF829 |
zinc finger protein 829 |
| chr19_-_37019562 | 0.17 |
ENST00000523638.1 |
ZNF260 |
zinc finger protein 260 |
| chr19_+_56905024 | 0.17 |
ENST00000591172.1 ENST00000589888.1 ENST00000587979.1 ENST00000585659.1 ENST00000593109.1 |
ZNF582-AS1 |
ZNF582 antisense RNA 1 (head to head) |
| chr1_+_220921640 | 0.17 |
ENST00000366913.3 |
MARC2 |
mitochondrial amidoxime reducing component 2 |
| chr16_-_279405 | 0.16 |
ENST00000430864.1 ENST00000293872.8 ENST00000337351.4 ENST00000397783.1 |
LUC7L |
LUC7-like (S. cerevisiae) |
| chr8_+_93895865 | 0.16 |
ENST00000391681.1 |
AC117834.1 |
AC117834.1 |
| chr1_+_53392901 | 0.16 |
ENST00000371514.3 ENST00000528311.1 ENST00000371509.4 ENST00000407246.2 ENST00000371513.5 |
SCP2 |
sterol carrier protein 2 |
| chr19_+_57050317 | 0.16 |
ENST00000301318.3 ENST00000591844.1 |
ZFP28 |
ZFP28 zinc finger protein |
| chr19_+_37341752 | 0.16 |
ENST00000586933.1 ENST00000532141.1 ENST00000420450.1 ENST00000526123.1 |
ZNF345 |
zinc finger protein 345 |
| chr17_-_18585541 | 0.16 |
ENST00000285274.5 ENST00000545289.1 ENST00000580145.1 |
ZNF286B |
zinc finger protein 286B |
| chr11_-_121986923 | 0.15 |
ENST00000560104.1 |
BLID |
BH3-like motif containing, cell death inducer |
| chr7_-_134001663 | 0.15 |
ENST00000378509.4 |
SLC35B4 |
solute carrier family 35 (UDP-xylose/UDP-N-acetylglucosamine transporter), member B4 |
| chr1_-_54519067 | 0.15 |
ENST00000452421.1 ENST00000420738.1 ENST00000234831.5 ENST00000440019.1 |
TMEM59 |
transmembrane protein 59 |
| chrX_-_70474910 | 0.15 |
ENST00000373988.1 ENST00000373998.1 |
ZMYM3 |
zinc finger, MYM-type 3 |
| chr17_+_32582293 | 0.15 |
ENST00000580907.1 ENST00000225831.4 |
CCL2 |
chemokine (C-C motif) ligand 2 |
| chr19_+_45445524 | 0.15 |
ENST00000591600.1 |
APOC4 |
apolipoprotein C-IV |
| chr12_-_10324716 | 0.14 |
ENST00000545927.1 ENST00000432556.2 ENST00000309539.3 ENST00000544577.1 |
OLR1 |
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr4_+_83956312 | 0.14 |
ENST00000509317.1 ENST00000503682.1 ENST00000511653.1 |
COPS4 |
COP9 signalosome subunit 4 |
| chr1_-_112106578 | 0.13 |
ENST00000369717.4 |
ADORA3 |
adenosine A3 receptor |
| chr6_+_2245982 | 0.12 |
ENST00000530346.1 ENST00000524770.1 ENST00000532124.1 ENST00000531092.1 ENST00000456943.2 ENST00000529893.1 |
GMDS-AS1 |
GMDS antisense RNA 1 (head to head) |
| chr12_+_80603233 | 0.12 |
ENST00000547103.1 ENST00000458043.2 |
OTOGL |
otogelin-like |
| chr19_+_56368803 | 0.12 |
ENST00000587891.1 |
NLRP4 |
NLR family, pyrin domain containing 4 |
| chr6_-_30658745 | 0.12 |
ENST00000376420.5 ENST00000376421.5 |
NRM |
nurim (nuclear envelope membrane protein) |
| chr2_-_178753465 | 0.12 |
ENST00000389683.3 |
PDE11A |
phosphodiesterase 11A |
| chr3_-_119379427 | 0.11 |
ENST00000264231.3 ENST00000468801.1 ENST00000538678.1 |
POPDC2 |
popeye domain containing 2 |
| chr6_+_88182643 | 0.11 |
ENST00000369556.3 ENST00000544441.1 ENST00000369552.4 ENST00000369557.5 |
SLC35A1 |
solute carrier family 35 (CMP-sialic acid transporter), member A1 |
| chr8_+_27632047 | 0.11 |
ENST00000397418.2 |
ESCO2 |
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr17_+_77021702 | 0.11 |
ENST00000392445.2 ENST00000354124.3 |
C1QTNF1 |
C1q and tumor necrosis factor related protein 1 |
| chr19_-_39881777 | 0.11 |
ENST00000595564.1 ENST00000221265.3 |
PAF1 |
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chr1_+_235530675 | 0.10 |
ENST00000366601.3 ENST00000406207.1 ENST00000543662.1 |
TBCE |
tubulin folding cofactor E |
| chr1_-_155112883 | 0.10 |
ENST00000368399.1 ENST00000368400.4 ENST00000341298.3 |
DPM3 |
dolichyl-phosphate mannosyltransferase polypeptide 3 |
| chr19_-_38183201 | 0.10 |
ENST00000590008.1 ENST00000358582.4 |
ZNF781 |
zinc finger protein 781 |
| chr19_+_44331493 | 0.10 |
ENST00000588797.1 |
ZNF283 |
zinc finger protein 283 |
| chr12_+_110011571 | 0.10 |
ENST00000539696.1 ENST00000228510.3 ENST00000392727.3 |
MVK |
mevalonate kinase |
| chr2_+_240684554 | 0.09 |
ENST00000407524.1 |
AC093802.1 |
Uncharacterized protein |
| chr1_-_16763685 | 0.09 |
ENST00000540400.1 |
SPATA21 |
spermatogenesis associated 21 |
| chr6_+_30131318 | 0.09 |
ENST00000376688.1 |
TRIM15 |
tripartite motif containing 15 |
| chr19_+_58281014 | 0.09 |
ENST00000391702.3 ENST00000598885.1 ENST00000598183.1 ENST00000396154.2 ENST00000599802.1 ENST00000396150.4 |
ZNF586 |
zinc finger protein 586 |
| chr3_+_14166440 | 0.09 |
ENST00000306077.4 |
TMEM43 |
transmembrane protein 43 |
| chr17_+_15602891 | 0.09 |
ENST00000421016.1 ENST00000593105.1 ENST00000580259.1 ENST00000583566.1 ENST00000472486.1 ENST00000395894.2 ENST00000581529.1 ENST00000579694.1 ENST00000580393.1 ENST00000585194.1 ENST00000583031.1 ENST00000464847.2 |
ZNF286A |
Homo sapiens zinc finger protein 286A (ZNF286A), transcript variant 6, mRNA. |
| chr19_-_37019136 | 0.09 |
ENST00000592282.1 |
ZNF260 |
zinc finger protein 260 |
| chr19_+_17530838 | 0.08 |
ENST00000528659.1 ENST00000392702.2 ENST00000529939.1 |
MVB12A |
multivesicular body subunit 12A |
| chr12_+_82347498 | 0.08 |
ENST00000550506.1 |
RP11-362A1.1 |
RP11-362A1.1 |
| chr12_+_21284118 | 0.08 |
ENST00000256958.2 |
SLCO1B1 |
solute carrier organic anion transporter family, member 1B1 |
| chr6_+_139094657 | 0.08 |
ENST00000332797.6 |
CCDC28A |
coiled-coil domain containing 28A |
| chr4_+_40194609 | 0.08 |
ENST00000508513.1 |
RHOH |
ras homolog family member H |
| chr15_-_83736091 | 0.08 |
ENST00000261721.4 |
BTBD1 |
BTB (POZ) domain containing 1 |
| chr19_+_9251052 | 0.08 |
ENST00000247956.6 ENST00000360385.3 |
ZNF317 |
zinc finger protein 317 |
| chr16_+_70207686 | 0.08 |
ENST00000541793.2 ENST00000314151.8 ENST00000565806.1 ENST00000569347.2 ENST00000536907.2 |
CLEC18C |
C-type lectin domain family 18, member C |
| chr3_-_130745571 | 0.08 |
ENST00000514044.1 ENST00000264992.3 |
ASTE1 |
asteroid homolog 1 (Drosophila) |
| chr19_-_58427944 | 0.08 |
ENST00000312026.5 ENST00000536263.1 ENST00000598629.1 ENST00000595559.1 ENST00000597515.1 ENST00000599251.1 ENST00000598526.1 |
ZNF417 |
zinc finger protein 417 |
| chr19_+_54041333 | 0.08 |
ENST00000411977.2 ENST00000511154.1 |
ZNF331 |
zinc finger protein 331 |
| chr19_+_49404041 | 0.08 |
ENST00000263273.5 ENST00000424608.1 |
NUCB1 |
nucleobindin 1 |
| chr19_-_39924349 | 0.07 |
ENST00000602153.1 |
RPS16 |
ribosomal protein S16 |
| chr19_-_913160 | 0.07 |
ENST00000361574.5 ENST00000587975.1 |
R3HDM4 |
R3H domain containing 4 |
| chr7_-_108096822 | 0.07 |
ENST00000379028.3 ENST00000413765.2 ENST00000379022.4 |
NRCAM |
neuronal cell adhesion molecule |
| chr7_+_142960505 | 0.07 |
ENST00000409500.3 ENST00000443571.2 ENST00000358406.5 ENST00000479303.1 |
GSTK1 |
glutathione S-transferase kappa 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.1 | 0.3 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.1 | 0.5 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.1 | 0.3 | GO:1902824 | positive regulation of late endosome to lysosome transport(GO:1902824) |
| 0.1 | 0.5 | GO:0090521 | regulation of embryonic cell shape(GO:0016476) septin cytoskeleton organization(GO:0032185) glomerular visceral epithelial cell migration(GO:0090521) |
| 0.1 | 0.3 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.1 | 1.7 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.1 | 1.4 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.1 | 0.3 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.1 | 0.2 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.1 | 1.0 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.1 | 0.2 | GO:1990569 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.1 | 0.2 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.0 | 0.3 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.4 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.3 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.3 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.3 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.2 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.1 | GO:0046373 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.0 | 0.5 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.2 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.0 | 0.3 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.5 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.1 | GO:0019075 | virus maturation(GO:0019075) |
| 0.0 | 0.5 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.3 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.5 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.4 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.0 | 0.6 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.3 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.1 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.2 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.2 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.2 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.1 | GO:0018317 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.0 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.0 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.0 | 0.1 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) terminal web assembly(GO:1902896) |
| 0.0 | 0.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.1 | 0.2 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.1 | 0.3 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.6 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.5 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 2.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.6 | GO:0044447 | axoneme part(GO:0044447) |
| 0.0 | 0.4 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.3 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.3 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.5 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.0 | GO:1990742 | microvesicle(GO:1990742) |
| 0.0 | 0.3 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.3 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.1 | 0.4 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.1 | 0.3 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.1 | 0.3 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 0.2 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.1 | 0.3 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.1 | 0.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 0.2 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.1 | 0.2 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.0 | 0.2 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.2 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.5 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.5 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 1.1 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.5 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.1 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 0.4 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.8 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.2 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.3 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.5 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 1.7 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.1 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.3 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.0 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.2 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 18.0 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.5 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.4 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.3 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.4 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |