Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for TBX19
Z-value: 0.53
Transcription factors associated with TBX19
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
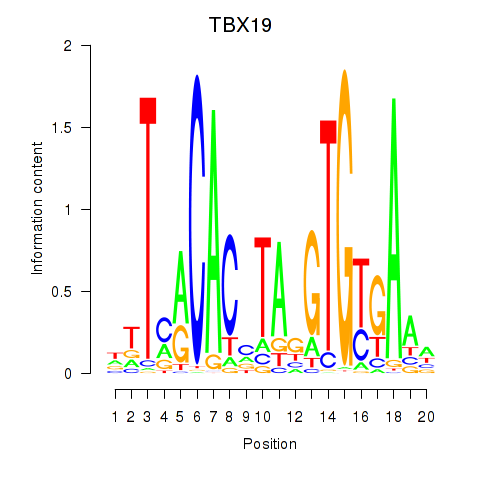
TBX19
|
ENSG00000143178.8 | TBX19 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TBX19 | hg19_v2_chr1_+_168250194_168250278 | 0.09 | 8.3e-01 | Click! |
Activity profile of TBX19 motif
Sorted Z-values of TBX19 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TBX19
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_113594279 | 0.90 |
ENST00000416750.1 ENST00000418817.1 ENST00000263341.2 |
IL1B |
interleukin 1, beta |
| chr1_+_46640750 | 0.79 |
ENST00000372003.1 |
TSPAN1 |
tetraspanin 1 |
| chr18_+_61554932 | 0.78 |
ENST00000299502.4 ENST00000457692.1 ENST00000413956.1 |
SERPINB2 |
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
| chr19_+_38755042 | 0.63 |
ENST00000301244.7 |
SPINT2 |
serine peptidase inhibitor, Kunitz type, 2 |
| chr18_-_47376197 | 0.52 |
ENST00000592688.1 |
MYO5B |
myosin VB |
| chr5_+_140165876 | 0.50 |
ENST00000504120.2 ENST00000394633.3 ENST00000378133.3 |
PCDHA1 |
protocadherin alpha 1 |
| chr11_-_119993734 | 0.50 |
ENST00000533302.1 |
TRIM29 |
tripartite motif containing 29 |
| chr6_-_2842087 | 0.49 |
ENST00000537185.1 |
SERPINB1 |
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr14_-_54420133 | 0.46 |
ENST00000559501.1 ENST00000558984.1 |
BMP4 |
bone morphogenetic protein 4 |
| chr10_+_95653687 | 0.41 |
ENST00000371408.3 ENST00000427197.1 |
SLC35G1 |
solute carrier family 35, member G1 |
| chr22_+_45072925 | 0.40 |
ENST00000006251.7 |
PRR5 |
proline rich 5 (renal) |
| chr1_-_153283194 | 0.40 |
ENST00000290722.1 |
PGLYRP3 |
peptidoglycan recognition protein 3 |
| chr5_-_33984786 | 0.40 |
ENST00000296589.4 |
SLC45A2 |
solute carrier family 45, member 2 |
| chr6_-_2842219 | 0.39 |
ENST00000380739.5 |
SERPINB1 |
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr3_+_98482175 | 0.38 |
ENST00000485391.1 ENST00000492254.1 |
ST3GAL6 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr22_+_45072958 | 0.38 |
ENST00000403581.1 |
PRR5 |
proline rich 5 (renal) |
| chr7_-_142139783 | 0.36 |
ENST00000390374.3 |
TRBV7-6 |
T cell receptor beta variable 7-6 |
| chr11_-_115375107 | 0.34 |
ENST00000545380.1 ENST00000452722.3 ENST00000537058.1 ENST00000536727.1 ENST00000542447.2 ENST00000331581.6 |
CADM1 |
cell adhesion molecule 1 |
| chr1_-_167522982 | 0.33 |
ENST00000370509.4 |
CREG1 |
cellular repressor of E1A-stimulated genes 1 |
| chr10_+_71561630 | 0.32 |
ENST00000398974.3 ENST00000398971.3 ENST00000398968.3 ENST00000398966.3 ENST00000398964.3 ENST00000398969.3 ENST00000356340.3 ENST00000398972.3 ENST00000398973.3 |
COL13A1 |
collagen, type XIII, alpha 1 |
| chr11_-_11747257 | 0.32 |
ENST00000601641.1 |
AC131935.1 |
AC131935.1 |
| chr3_+_118905564 | 0.29 |
ENST00000460625.1 |
UPK1B |
uroplakin 1B |
| chr20_-_22565101 | 0.28 |
ENST00000419308.2 |
FOXA2 |
forkhead box A2 |
| chr11_-_59383617 | 0.28 |
ENST00000263847.1 |
OSBP |
oxysterol binding protein |
| chr19_+_46732988 | 0.25 |
ENST00000437936.1 |
IGFL1 |
IGF-like family member 1 |
| chr12_-_9360966 | 0.24 |
ENST00000261336.2 |
PZP |
pregnancy-zone protein |
| chr3_+_57741957 | 0.24 |
ENST00000295951.3 |
SLMAP |
sarcolemma associated protein |
| chr17_+_55162453 | 0.21 |
ENST00000575322.1 ENST00000337714.3 ENST00000314126.3 |
AKAP1 |
A kinase (PRKA) anchor protein 1 |
| chr6_-_26216872 | 0.20 |
ENST00000244601.3 |
HIST1H2BG |
histone cluster 1, H2bg |
| chr1_+_172389821 | 0.19 |
ENST00000367727.4 |
C1orf105 |
chromosome 1 open reading frame 105 |
| chr14_+_70346125 | 0.19 |
ENST00000361956.3 ENST00000381280.4 |
SMOC1 |
SPARC related modular calcium binding 1 |
| chr1_-_116311402 | 0.18 |
ENST00000261448.5 |
CASQ2 |
calsequestrin 2 (cardiac muscle) |
| chr14_+_23727694 | 0.18 |
ENST00000399905.1 ENST00000470456.1 |
C14orf164 |
chromosome 14 open reading frame 164 |
| chr1_-_100598444 | 0.17 |
ENST00000535161.1 ENST00000287482.5 |
SASS6 |
spindle assembly 6 homolog (C. elegans) |
| chr5_-_78281775 | 0.17 |
ENST00000396151.3 ENST00000565165.1 |
ARSB |
arylsulfatase B |
| chr8_+_125954281 | 0.17 |
ENST00000510897.2 ENST00000533286.1 |
LINC00964 |
long intergenic non-protein coding RNA 964 |
| chr14_+_32546274 | 0.17 |
ENST00000396582.2 |
ARHGAP5 |
Rho GTPase activating protein 5 |
| chr5_-_33984741 | 0.16 |
ENST00000382102.3 ENST00000509381.1 ENST00000342059.3 ENST00000345083.5 |
SLC45A2 |
solute carrier family 45, member 2 |
| chr3_+_94657086 | 0.15 |
ENST00000463200.1 |
LINC00879 |
long intergenic non-protein coding RNA 879 |
| chr14_+_32546145 | 0.15 |
ENST00000556611.1 ENST00000539826.2 |
ARHGAP5 |
Rho GTPase activating protein 5 |
| chr6_-_133035185 | 0.15 |
ENST00000367928.4 |
VNN1 |
vanin 1 |
| chr11_-_5462744 | 0.14 |
ENST00000380211.1 |
OR51I1 |
olfactory receptor, family 51, subfamily I, member 1 |
| chr12_-_10962767 | 0.14 |
ENST00000240691.2 |
TAS2R9 |
taste receptor, type 2, member 9 |
| chr2_+_201754135 | 0.14 |
ENST00000409357.1 ENST00000409129.2 |
NIF3L1 |
NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae) |
| chr1_+_207627575 | 0.14 |
ENST00000367058.3 ENST00000367057.3 ENST00000367059.3 |
CR2 |
complement component (3d/Epstein Barr virus) receptor 2 |
| chr2_+_201754050 | 0.14 |
ENST00000426253.1 ENST00000416651.1 ENST00000454952.1 ENST00000409020.1 ENST00000359683.4 |
NIF3L1 |
NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae) |
| chr6_-_117086873 | 0.14 |
ENST00000368557.4 |
FAM162B |
family with sequence similarity 162, member B |
| chr12_-_11175219 | 0.14 |
ENST00000390673.2 |
TAS2R19 |
taste receptor, type 2, member 19 |
| chr7_+_2394445 | 0.13 |
ENST00000360876.4 ENST00000413917.1 ENST00000397011.2 |
EIF3B |
eukaryotic translation initiation factor 3, subunit B |
| chr16_+_84801852 | 0.12 |
ENST00000569925.1 ENST00000567526.1 |
USP10 |
ubiquitin specific peptidase 10 |
| chr8_-_6836156 | 0.12 |
ENST00000382679.2 |
DEFA1 |
defensin, alpha 1 |
| chr1_+_16090914 | 0.12 |
ENST00000441801.2 |
FBLIM1 |
filamin binding LIM protein 1 |
| chrX_+_107288239 | 0.12 |
ENST00000217957.5 |
VSIG1 |
V-set and immunoglobulin domain containing 1 |
| chr4_-_156298028 | 0.11 |
ENST00000433024.1 ENST00000379248.2 |
MAP9 |
microtubule-associated protein 9 |
| chr12_-_8218997 | 0.11 |
ENST00000307637.4 |
C3AR1 |
complement component 3a receptor 1 |
| chr11_+_46740730 | 0.11 |
ENST00000311907.5 ENST00000530231.1 ENST00000442468.1 |
F2 |
coagulation factor II (thrombin) |
| chr8_+_94767072 | 0.11 |
ENST00000452276.1 ENST00000453321.3 ENST00000498673.1 ENST00000518319.1 |
TMEM67 |
transmembrane protein 67 |
| chr20_-_50385138 | 0.11 |
ENST00000338821.5 |
ATP9A |
ATPase, class II, type 9A |
| chr7_-_43946175 | 0.11 |
ENST00000603700.1 ENST00000402306.3 ENST00000443736.1 ENST00000446958.1 |
URGCP-MRPS24 URGCP |
URGCP-MRPS24 readthrough upregulator of cell proliferation |
| chr19_-_39421377 | 0.11 |
ENST00000430193.3 ENST00000600042.1 ENST00000221431.6 |
SARS2 |
seryl-tRNA synthetase 2, mitochondrial |
| chr22_+_31477296 | 0.11 |
ENST00000426927.1 ENST00000440425.1 ENST00000358743.1 ENST00000347557.2 ENST00000333137.7 |
SMTN |
smoothelin |
| chr7_+_93535817 | 0.11 |
ENST00000248572.5 |
GNGT1 |
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr14_+_69658194 | 0.10 |
ENST00000409018.3 ENST00000409014.1 ENST00000409675.1 |
EXD2 |
exonuclease 3'-5' domain containing 2 |
| chr2_+_90153696 | 0.10 |
ENST00000417279.2 |
IGKV3D-15 |
immunoglobulin kappa variable 3D-15 (gene/pseudogene) |
| chr6_+_26217159 | 0.10 |
ENST00000303910.2 |
HIST1H2AE |
histone cluster 1, H2ae |
| chr17_+_4736627 | 0.10 |
ENST00000355280.6 ENST00000347992.7 |
MINK1 |
misshapen-like kinase 1 |
| chr7_+_93535866 | 0.10 |
ENST00000429473.1 ENST00000430875.1 ENST00000428834.1 |
GNGT1 |
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr16_+_66995121 | 0.10 |
ENST00000303334.4 |
CES3 |
carboxylesterase 3 |
| chr7_-_112430427 | 0.09 |
ENST00000449743.1 ENST00000441474.1 ENST00000454074.1 ENST00000447395.1 |
TMEM168 |
transmembrane protein 168 |
| chr16_+_66995144 | 0.09 |
ENST00000394037.1 |
CES3 |
carboxylesterase 3 |
| chr11_+_34645791 | 0.09 |
ENST00000529527.1 ENST00000531728.1 ENST00000525253.1 |
EHF |
ets homologous factor |
| chr1_+_160765947 | 0.09 |
ENST00000263285.6 ENST00000368039.2 |
LY9 |
lymphocyte antigen 9 |
| chr2_+_228337079 | 0.09 |
ENST00000409315.1 ENST00000373671.3 ENST00000409171.1 |
AGFG1 |
ArfGAP with FG repeats 1 |
| chr12_+_120740119 | 0.09 |
ENST00000536460.1 ENST00000202967.4 |
SIRT4 |
sirtuin 4 |
| chr16_-_2031464 | 0.09 |
ENST00000356120.4 ENST00000354249.4 |
NOXO1 |
NADPH oxidase organizer 1 |
| chr20_-_44991813 | 0.09 |
ENST00000372227.1 |
SLC35C2 |
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr2_-_64751227 | 0.09 |
ENST00000561559.1 |
RP11-568N6.1 |
RP11-568N6.1 |
| chr6_-_10694766 | 0.09 |
ENST00000460742.2 ENST00000259983.3 ENST00000379586.1 |
C6orf52 |
chromosome 6 open reading frame 52 |
| chr6_+_26158343 | 0.09 |
ENST00000377777.4 ENST00000289316.2 |
HIST1H2BD |
histone cluster 1, H2bd |
| chr14_+_105147464 | 0.09 |
ENST00000540171.2 |
RP11-982M15.6 |
RP11-982M15.6 |
| chrX_+_107288197 | 0.08 |
ENST00000415430.3 |
VSIG1 |
V-set and immunoglobulin domain containing 1 |
| chr17_-_34207295 | 0.08 |
ENST00000463941.1 ENST00000293272.3 |
CCL5 |
chemokine (C-C motif) ligand 5 |
| chr6_+_123110465 | 0.08 |
ENST00000539041.1 |
SMPDL3A |
sphingomyelin phosphodiesterase, acid-like 3A |
| chr15_-_64338521 | 0.08 |
ENST00000457488.1 ENST00000558069.1 |
DAPK2 |
death-associated protein kinase 2 |
| chr19_+_46003056 | 0.08 |
ENST00000401593.1 ENST00000396736.2 |
PPM1N |
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr2_-_89278535 | 0.08 |
ENST00000390247.2 |
IGKV3-7 |
immunoglobulin kappa variable 3-7 (non-functional) |
| chr5_-_93077293 | 0.08 |
ENST00000510627.4 |
POU5F2 |
POU domain class 5, transcription factor 2 |
| chr17_+_41561317 | 0.08 |
ENST00000540306.1 ENST00000262415.3 ENST00000605777.1 |
DHX8 |
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chr8_+_94767109 | 0.07 |
ENST00000409623.3 ENST00000453906.1 ENST00000521517.1 |
TMEM67 |
transmembrane protein 67 |
| chr7_+_134671234 | 0.07 |
ENST00000436302.2 ENST00000359383.3 ENST00000458078.1 ENST00000435976.2 ENST00000455283.2 |
AGBL3 |
ATP/GTP binding protein-like 3 |
| chr14_+_51706886 | 0.07 |
ENST00000457354.2 |
TMX1 |
thioredoxin-related transmembrane protein 1 |
| chr7_+_47834880 | 0.07 |
ENST00000258776.4 |
C7orf69 |
chromosome 7 open reading frame 69 |
| chr19_-_36297632 | 0.07 |
ENST00000588266.2 |
PRODH2 |
proline dehydrogenase (oxidase) 2 |
| chrX_-_138914394 | 0.07 |
ENST00000327569.3 ENST00000361648.2 ENST00000370543.1 ENST00000359686.2 |
ATP11C |
ATPase, class VI, type 11C |
| chr15_+_43425672 | 0.07 |
ENST00000260403.2 |
TMEM62 |
transmembrane protein 62 |
| chr3_-_194393206 | 0.07 |
ENST00000265245.5 |
LSG1 |
large 60S subunit nuclear export GTPase 1 |
| chr14_+_21249200 | 0.07 |
ENST00000304677.2 |
RNASE6 |
ribonuclease, RNase A family, k6 |
| chr9_+_130478345 | 0.06 |
ENST00000373289.3 ENST00000393748.4 |
TTC16 |
tetratricopeptide repeat domain 16 |
| chr17_+_35306175 | 0.06 |
ENST00000225402.5 |
AATF |
apoptosis antagonizing transcription factor |
| chr19_-_8675559 | 0.06 |
ENST00000597188.1 |
ADAMTS10 |
ADAM metallopeptidase with thrombospondin type 1 motif, 10 |
| chr3_+_132316081 | 0.06 |
ENST00000249887.2 |
ACKR4 |
atypical chemokine receptor 4 |
| chr18_-_3874752 | 0.06 |
ENST00000534970.1 |
DLGAP1 |
discs, large (Drosophila) homolog-associated protein 1 |
| chr3_+_46204959 | 0.06 |
ENST00000357422.2 |
CCR3 |
chemokine (C-C motif) receptor 3 |
| chr6_+_10694900 | 0.06 |
ENST00000379568.3 |
PAK1IP1 |
PAK1 interacting protein 1 |
| chr3_+_35722487 | 0.06 |
ENST00000441454.1 |
ARPP21 |
cAMP-regulated phosphoprotein, 21kDa |
| chr12_-_49319265 | 0.06 |
ENST00000552878.1 ENST00000453172.2 |
FKBP11 |
FK506 binding protein 11, 19 kDa |
| chr10_+_12171636 | 0.06 |
ENST00000379051.1 ENST00000379033.3 ENST00000441368.1 ENST00000298428.9 ENST00000304267.8 |
SEC61A2 |
Sec61 alpha 2 subunit (S. cerevisiae) |
| chr7_+_112120908 | 0.06 |
ENST00000439068.2 ENST00000312849.4 ENST00000429049.1 |
LSMEM1 |
leucine-rich single-pass membrane protein 1 |
| chr14_+_69658480 | 0.05 |
ENST00000409949.1 ENST00000409242.1 ENST00000312994.5 ENST00000413191.1 |
EXD2 |
exonuclease 3'-5' domain containing 2 |
| chr16_+_19222479 | 0.05 |
ENST00000568433.1 |
SYT17 |
synaptotagmin XVII |
| chr10_-_12237820 | 0.05 |
ENST00000378937.3 ENST00000378927.3 |
NUDT5 |
nudix (nucleoside diphosphate linked moiety X)-type motif 5 |
| chr18_+_56892724 | 0.05 |
ENST00000456142.3 ENST00000530323.1 |
GRP |
gastrin-releasing peptide |
| chr1_+_96457545 | 0.05 |
ENST00000413825.2 |
RP11-147C23.1 |
Uncharacterized protein |
| chr5_+_96211643 | 0.05 |
ENST00000437043.3 ENST00000510373.1 |
ERAP2 |
endoplasmic reticulum aminopeptidase 2 |
| chrX_-_15402498 | 0.05 |
ENST00000297904.3 |
FIGF |
c-fos induced growth factor (vascular endothelial growth factor D) |
| chr11_-_84028180 | 0.05 |
ENST00000280241.8 |
DLG2 |
discs, large homolog 2 (Drosophila) |
| chr10_-_12237836 | 0.05 |
ENST00000444732.1 ENST00000378940.3 |
NUDT5 |
nudix (nucleoside diphosphate linked moiety X)-type motif 5 |
| chr12_-_9760482 | 0.05 |
ENST00000229402.3 |
KLRB1 |
killer cell lectin-like receptor subfamily B, member 1 |
| chr16_-_20339123 | 0.05 |
ENST00000381360.5 |
GP2 |
glycoprotein 2 (zymogen granule membrane) |
| chr9_-_100684845 | 0.05 |
ENST00000375119.3 |
C9orf156 |
chromosome 9 open reading frame 156 |
| chr7_-_151137094 | 0.04 |
ENST00000491928.1 ENST00000337323.2 |
CRYGN |
crystallin, gamma N |
| chr9_+_78505581 | 0.04 |
ENST00000376767.3 ENST00000376752.4 |
PCSK5 |
proprotein convertase subtilisin/kexin type 5 |
| chr5_+_33440802 | 0.04 |
ENST00000502553.1 ENST00000514259.1 ENST00000265112.3 |
TARS |
threonyl-tRNA synthetase |
| chr11_+_5474638 | 0.04 |
ENST00000341449.2 |
OR51I2 |
olfactory receptor, family 51, subfamily I, member 2 |
| chr4_-_4249924 | 0.04 |
ENST00000254742.2 ENST00000382753.4 ENST00000540397.1 ENST00000538516.1 |
TMEM128 |
transmembrane protein 128 |
| chr6_+_158957431 | 0.04 |
ENST00000367090.3 |
TMEM181 |
transmembrane protein 181 |
| chr1_+_153963227 | 0.04 |
ENST00000368567.4 ENST00000392558.4 |
RPS27 |
ribosomal protein S27 |
| chr3_+_51863433 | 0.04 |
ENST00000444293.1 |
IQCF3 |
IQ motif containing F3 |
| chr1_+_100598691 | 0.04 |
ENST00000370143.1 ENST00000370141.2 |
TRMT13 |
tRNA methyltransferase 13 homolog (S. cerevisiae) |
| chr1_+_236557569 | 0.04 |
ENST00000334232.4 |
EDARADD |
EDAR-associated death domain |
| chr11_+_118398178 | 0.04 |
ENST00000302783.4 ENST00000539546.1 |
TTC36 |
tetratricopeptide repeat domain 36 |
| chr5_+_96212185 | 0.04 |
ENST00000379904.4 |
ERAP2 |
endoplasmic reticulum aminopeptidase 2 |
| chr15_+_84908573 | 0.04 |
ENST00000424966.1 ENST00000422563.2 |
GOLGA6L4 |
golgin A6 family-like 4 |
| chr1_-_154458520 | 0.04 |
ENST00000486773.1 |
SHE |
Src homology 2 domain containing E |
| chr5_-_179072047 | 0.04 |
ENST00000448248.2 |
C5orf60 |
chromosome 5 open reading frame 60 |
| chr19_-_15575369 | 0.04 |
ENST00000343625.7 |
RASAL3 |
RAS protein activator like 3 |
| chr6_+_147091575 | 0.03 |
ENST00000326916.8 ENST00000470716.2 ENST00000367488.1 |
ADGB |
androglobin |
| chr1_-_100715372 | 0.03 |
ENST00000370131.3 ENST00000370132.4 |
DBT |
dihydrolipoamide branched chain transacylase E2 |
| chr5_-_94417339 | 0.03 |
ENST00000429576.2 ENST00000508509.1 ENST00000510732.1 |
MCTP1 |
multiple C2 domains, transmembrane 1 |
| chr19_+_7049332 | 0.03 |
ENST00000381393.3 |
MBD3L2 |
methyl-CpG binding domain protein 3-like 2 |
| chr9_+_78505554 | 0.03 |
ENST00000545128.1 |
PCSK5 |
proprotein convertase subtilisin/kexin type 5 |
| chr11_-_19262486 | 0.03 |
ENST00000250024.4 |
E2F8 |
E2F transcription factor 8 |
| chr6_+_42847348 | 0.03 |
ENST00000493763.1 ENST00000304734.5 |
RPL7L1 |
ribosomal protein L7-like 1 |
| chr3_-_179169181 | 0.03 |
ENST00000497513.1 |
GNB4 |
guanine nucleotide binding protein (G protein), beta polypeptide 4 |
| chr2_-_217724767 | 0.03 |
ENST00000236979.2 |
TNP1 |
transition protein 1 (during histone to protamine replacement) |
| chr2_-_26864228 | 0.03 |
ENST00000288861.4 |
CIB4 |
calcium and integrin binding family member 4 |
| chr5_-_94417314 | 0.03 |
ENST00000505208.1 |
MCTP1 |
multiple C2 domains, transmembrane 1 |
| chr17_+_3118915 | 0.03 |
ENST00000304094.1 |
OR1A1 |
olfactory receptor, family 1, subfamily A, member 1 |
| chrX_+_79675965 | 0.03 |
ENST00000308293.5 |
FAM46D |
family with sequence similarity 46, member D |
| chr8_+_125463048 | 0.03 |
ENST00000328599.3 |
TRMT12 |
tRNA methyltransferase 12 homolog (S. cerevisiae) |
| chr1_+_117297007 | 0.03 |
ENST00000369478.3 ENST00000369477.1 |
CD2 |
CD2 molecule |
| chr20_+_44036620 | 0.03 |
ENST00000372710.3 |
DBNDD2 |
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr6_-_137366096 | 0.03 |
ENST00000316649.5 ENST00000367746.3 |
IL20RA |
interleukin 20 receptor, alpha |
| chr19_+_58111241 | 0.02 |
ENST00000597700.1 ENST00000332854.6 ENST00000597864.1 |
ZNF530 |
zinc finger protein 530 |
| chr11_+_57425209 | 0.02 |
ENST00000533905.1 ENST00000525602.1 ENST00000302731.4 |
CLP1 |
cleavage and polyadenylation factor I subunit 1 |
| chr16_-_2770216 | 0.02 |
ENST00000302641.3 |
PRSS27 |
protease, serine 27 |
| chr2_-_95787738 | 0.02 |
ENST00000272418.2 |
MRPS5 |
mitochondrial ribosomal protein S5 |
| chr1_-_150947343 | 0.02 |
ENST00000271688.6 ENST00000368954.5 |
CERS2 |
ceramide synthase 2 |
| chr2_+_62900986 | 0.02 |
ENST00000405015.3 ENST00000413434.1 ENST00000426940.1 ENST00000449820.1 |
EHBP1 |
EH domain binding protein 1 |
| chr19_+_49496705 | 0.02 |
ENST00000595090.1 |
RUVBL2 |
RuvB-like AAA ATPase 2 |
| chr9_+_116173000 | 0.02 |
ENST00000288462.4 |
C9orf43 |
chromosome 9 open reading frame 43 |
| chr22_-_31364187 | 0.02 |
ENST00000215862.4 ENST00000397641.3 |
MORC2 |
MORC family CW-type zinc finger 2 |
| chr6_-_117150198 | 0.02 |
ENST00000310357.3 ENST00000368549.3 ENST00000530250.1 |
GPRC6A |
G protein-coupled receptor, family C, group 6, member A |
| chr2_-_9563575 | 0.02 |
ENST00000488451.1 ENST00000238091.4 ENST00000355346.4 |
ITGB1BP1 |
integrin beta 1 binding protein 1 |
| chr19_+_49496782 | 0.02 |
ENST00000601968.1 ENST00000596837.1 |
RUVBL2 |
RuvB-like AAA ATPase 2 |
| chr12_-_11036844 | 0.02 |
ENST00000428168.2 |
PRH1 |
proline-rich protein HaeIII subfamily 1 |
| chr3_-_128185811 | 0.02 |
ENST00000469083.1 |
DNAJB8 |
DnaJ (Hsp40) homolog, subfamily B, member 8 |
| chrX_+_129535937 | 0.02 |
ENST00000305536.6 ENST00000370947.1 |
RBMX2 |
RNA binding motif protein, X-linked 2 |
| chr19_+_21203426 | 0.02 |
ENST00000261560.5 ENST00000599548.1 ENST00000594110.1 |
ZNF430 |
zinc finger protein 430 |
| chr11_-_59436453 | 0.01 |
ENST00000300146.9 |
PATL1 |
protein associated with topoisomerase II homolog 1 (yeast) |
| chr10_+_106034884 | 0.01 |
ENST00000369707.2 ENST00000429569.2 |
GSTO2 |
glutathione S-transferase omega 2 |
| chr3_+_119499331 | 0.01 |
ENST00000393716.2 ENST00000466380.1 |
NR1I2 |
nuclear receptor subfamily 1, group I, member 2 |
| chr6_+_42847649 | 0.01 |
ENST00000424341.2 ENST00000602561.1 |
RPL7L1 |
ribosomal protein L7-like 1 |
| chr10_-_120938303 | 0.01 |
ENST00000356951.3 ENST00000298510.2 |
PRDX3 |
peroxiredoxin 3 |
| chr22_-_47882857 | 0.01 |
ENST00000405369.3 |
LL22NC03-75H12.2 |
Novel protein; Uncharacterized protein |
| chr12_-_11214893 | 0.01 |
ENST00000533467.1 |
TAS2R46 |
taste receptor, type 2, member 46 |
| chr14_-_58894332 | 0.01 |
ENST00000395159.2 |
TIMM9 |
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr15_-_65809581 | 0.01 |
ENST00000341861.5 |
DPP8 |
dipeptidyl-peptidase 8 |
| chr3_-_150264272 | 0.01 |
ENST00000491660.1 ENST00000487153.1 ENST00000239944.2 |
SERP1 |
stress-associated endoplasmic reticulum protein 1 |
| chr9_+_116172958 | 0.01 |
ENST00000374165.1 |
C9orf43 |
chromosome 9 open reading frame 43 |
| chr12_-_10875831 | 0.01 |
ENST00000279550.7 ENST00000228251.4 |
YBX3 |
Y box binding protein 3 |
| chr12_+_8276495 | 0.01 |
ENST00000546339.1 |
CLEC4A |
C-type lectin domain family 4, member A |
| chr15_+_26360970 | 0.01 |
ENST00000556159.1 ENST00000557523.1 |
LINC00929 |
long intergenic non-protein coding RNA 929 |
| chr3_+_32726620 | 0.00 |
ENST00000331889.6 ENST00000328834.5 |
CNOT10 |
CCR4-NOT transcription complex, subunit 10 |
| chr9_-_130890662 | 0.00 |
ENST00000277462.5 ENST00000338961.6 |
PTGES2 |
prostaglandin E synthase 2 |
| chr8_-_6795823 | 0.00 |
ENST00000297435.2 |
DEFA4 |
defensin, alpha 4, corticostatin |
| chr19_-_8008533 | 0.00 |
ENST00000597926.1 |
TIMM44 |
translocase of inner mitochondrial membrane 44 homolog (yeast) |
| chr8_-_134115118 | 0.00 |
ENST00000395352.3 ENST00000338087.5 |
SLA |
Src-like-adaptor |
| chr9_+_77230499 | 0.00 |
ENST00000396204.2 |
RORB |
RAR-related orphan receptor B |
| chr7_+_76751926 | 0.00 |
ENST00000285871.4 ENST00000431197.1 |
CCDC146 |
coiled-coil domain containing 146 |
| chr19_-_40331345 | 0.00 |
ENST00000597224.1 |
FBL |
fibrillarin |
| chr7_+_117251671 | 0.00 |
ENST00000468795.1 |
CFTR |
cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) |
| chr1_-_63988846 | 0.00 |
ENST00000283568.8 ENST00000371092.3 ENST00000271002.10 |
ITGB3BP |
integrin beta 3 binding protein (beta3-endonexin) |
| chr1_-_157567868 | 0.00 |
ENST00000271532.1 |
FCRL4 |
Fc receptor-like 4 |
| chr10_+_76969909 | 0.00 |
ENST00000298468.5 ENST00000543351.1 |
VDAC2 |
voltage-dependent anion channel 2 |
| chr10_+_12237924 | 0.00 |
ENST00000429258.2 ENST00000281141.4 |
CDC123 |
cell division cycle 123 |
| chr10_-_97200772 | 0.00 |
ENST00000371241.1 ENST00000354106.3 ENST00000371239.1 ENST00000361941.3 ENST00000277982.5 ENST00000371245.3 |
SORBS1 |
sorbin and SH3 domain containing 1 |
| chr19_+_535835 | 0.00 |
ENST00000607527.1 ENST00000606065.1 |
CDC34 |
cell division cycle 34 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0060557 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.2 | 0.6 | GO:0015766 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.2 | 0.5 | GO:0072096 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.1 | 0.6 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.1 | 0.3 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.1 | 0.3 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 | 0.2 | GO:0032824 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.1 | 0.5 | GO:0032439 | endosome localization(GO:0032439) |
| 0.1 | 0.8 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.4 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.1 | GO:0035915 | pore formation in membrane of other organism(GO:0035915) |
| 0.0 | 0.1 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.2 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.1 | GO:0061582 | intestinal epithelial cell migration(GO:0061582) |
| 0.0 | 0.1 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.8 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.1 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.0 | 0.1 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.3 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.2 | GO:0086068 | Purkinje myocyte to ventricular cardiac muscle cell signaling(GO:0086029) Purkinje myocyte to ventricular cardiac muscle cell communication(GO:0086068) |
| 0.0 | 0.1 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.0 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.1 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.1 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.1 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 | 0.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.4 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.0 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.0 | 0.0 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.2 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.0 | 0.5 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.8 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.3 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.1 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0015154 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 0.4 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.1 | 0.2 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 0.9 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.5 | GO:0070700 | co-receptor binding(GO:0039706) BMP receptor binding(GO:0070700) |
| 0.0 | 0.3 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.0 | 0.1 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.0 | 2.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.1 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.2 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.5 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.0 | 0.5 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.0 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.9 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |