Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for TBX20
Z-value: 0.67
Transcription factors associated with TBX20
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX20
|
ENSG00000164532.10 | TBX20 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TBX20 | hg19_v2_chr7_-_35293740_35293758 | -0.43 | 2.9e-01 | Click! |
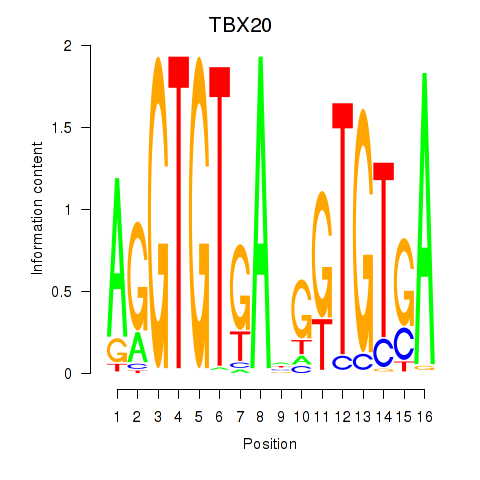
Activity profile of TBX20 motif
Sorted Z-values of TBX20 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TBX20
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_57662419 | 4.38 |
ENST00000388812.4 ENST00000538815.1 ENST00000456916.1 ENST00000567154.1 ENST00000388813.5 ENST00000562558.1 ENST00000566271.2 |
GPR56 |
G protein-coupled receptor 56 |
| chr17_-_3599492 | 0.99 |
ENST00000435558.1 ENST00000345901.3 |
P2RX5 |
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr6_-_136788001 | 0.99 |
ENST00000544465.1 |
MAP7 |
microtubule-associated protein 7 |
| chr17_-_3599327 | 0.98 |
ENST00000551178.1 ENST00000552276.1 ENST00000547178.1 |
P2RX5 |
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr2_+_85360499 | 0.88 |
ENST00000282111.3 |
TCF7L1 |
transcription factor 7-like 1 (T-cell specific, HMG-box) |
| chr17_-_3599696 | 0.80 |
ENST00000225328.5 |
P2RX5 |
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr6_-_2842087 | 0.67 |
ENST00000537185.1 |
SERPINB1 |
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr6_-_2842219 | 0.55 |
ENST00000380739.5 |
SERPINB1 |
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr7_-_139763521 | 0.46 |
ENST00000263549.3 |
PARP12 |
poly (ADP-ribose) polymerase family, member 12 |
| chr2_+_207630081 | 0.31 |
ENST00000236980.6 ENST00000418289.1 ENST00000402774.3 ENST00000403094.3 |
FASTKD2 |
FAST kinase domains 2 |
| chr3_+_142315225 | 0.31 |
ENST00000457734.2 ENST00000483373.1 ENST00000475296.1 ENST00000495744.1 ENST00000476044.1 ENST00000461644.1 |
PLS1 |
plastin 1 |
| chr10_-_131762105 | 0.23 |
ENST00000368648.3 ENST00000355311.5 |
EBF3 |
early B-cell factor 3 |
| chr2_+_61108650 | 0.23 |
ENST00000295025.8 |
REL |
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr1_-_158300747 | 0.22 |
ENST00000451207.1 |
CD1B |
CD1b molecule |
| chr5_+_135394840 | 0.22 |
ENST00000503087.1 |
TGFBI |
transforming growth factor, beta-induced, 68kDa |
| chr2_+_61108771 | 0.21 |
ENST00000394479.3 |
REL |
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr3_+_40428647 | 0.21 |
ENST00000301825.3 ENST00000439533.1 ENST00000456402.1 |
ENTPD3 |
ectonucleoside triphosphate diphosphohydrolase 3 |
| chr5_+_161494521 | 0.21 |
ENST00000356592.3 |
GABRG2 |
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr2_+_176972000 | 0.18 |
ENST00000249504.5 |
HOXD11 |
homeobox D11 |
| chr15_-_22448819 | 0.16 |
ENST00000604066.1 |
IGHV1OR15-1 |
immunoglobulin heavy variable 1/OR15-1 (non-functional) |
| chr3_-_139396853 | 0.16 |
ENST00000406164.1 ENST00000406824.1 |
NMNAT3 |
nicotinamide nucleotide adenylyltransferase 3 |
| chr1_+_22964073 | 0.14 |
ENST00000402322.1 |
C1QA |
complement component 1, q subcomponent, A chain |
| chr3_-_139396801 | 0.14 |
ENST00000413939.2 ENST00000339837.5 ENST00000512391.1 |
NMNAT3 |
nicotinamide nucleotide adenylyltransferase 3 |
| chr3_-_50396978 | 0.13 |
ENST00000266025.3 |
TMEM115 |
transmembrane protein 115 |
| chr11_+_6281036 | 0.13 |
ENST00000532715.1 ENST00000525014.1 ENST00000531712.1 ENST00000525462.1 |
CCKBR |
cholecystokinin B receptor |
| chr1_+_43824669 | 0.12 |
ENST00000372462.1 |
CDC20 |
cell division cycle 20 |
| chr10_+_114135004 | 0.12 |
ENST00000393081.1 |
ACSL5 |
acyl-CoA synthetase long-chain family member 5 |
| chr1_-_11865982 | 0.12 |
ENST00000418034.1 |
MTHFR |
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr20_+_31805131 | 0.11 |
ENST00000375454.3 ENST00000375452.3 |
BPIFA3 |
BPI fold containing family A, member 3 |
| chr6_-_107077347 | 0.10 |
ENST00000369063.3 ENST00000539449.1 |
RTN4IP1 |
reticulon 4 interacting protein 1 |
| chrX_+_7810303 | 0.09 |
ENST00000381059.3 ENST00000341408.4 |
VCX |
variable charge, X-linked |
| chr1_-_11118896 | 0.09 |
ENST00000465788.1 |
SRM |
spermidine synthase |
| chr10_+_114135952 | 0.09 |
ENST00000356116.1 ENST00000433418.1 ENST00000354273.4 |
ACSL5 |
acyl-CoA synthetase long-chain family member 5 |
| chr2_-_219134822 | 0.09 |
ENST00000444053.1 ENST00000248450.4 |
AAMP |
angio-associated, migratory cell protein |
| chr7_-_142099977 | 0.09 |
ENST00000390359.3 |
TRBV7-8 |
T cell receptor beta variable 7-8 |
| chrX_-_52386980 | 0.08 |
ENST00000330906.4 |
XAGE2 |
X antigen family, member 2 |
| chr14_-_106453155 | 0.08 |
ENST00000390594.2 |
IGHV1-2 |
immunoglobulin heavy variable 1-2 |
| chr1_+_87797351 | 0.08 |
ENST00000370542.1 |
LMO4 |
LIM domain only 4 |
| chr19_+_44331493 | 0.07 |
ENST00000588797.1 |
ZNF283 |
zinc finger protein 283 |
| chrX_+_52112158 | 0.07 |
ENST00000286049.2 |
XAGE2B |
X antigen family, member 2B |
| chr12_-_23737534 | 0.06 |
ENST00000396007.2 |
SOX5 |
SRY (sex determining region Y)-box 5 |
| chr10_+_88414298 | 0.05 |
ENST00000372071.2 |
OPN4 |
opsin 4 |
| chr10_+_88414338 | 0.05 |
ENST00000241891.5 ENST00000443292.1 |
OPN4 |
opsin 4 |
| chrX_+_67913471 | 0.04 |
ENST00000374597.3 |
STARD8 |
StAR-related lipid transfer (START) domain containing 8 |
| chr1_+_147013182 | 0.04 |
ENST00000234739.3 |
BCL9 |
B-cell CLL/lymphoma 9 |
| chr3_-_131221790 | 0.03 |
ENST00000512877.1 ENST00000264995.3 ENST00000511168.1 ENST00000425847.2 |
MRPL3 |
mitochondrial ribosomal protein L3 |
| chr1_-_17380630 | 0.02 |
ENST00000375499.3 |
SDHB |
succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
| chr15_-_38519066 | 0.02 |
ENST00000561320.1 ENST00000561161.1 |
RP11-346D14.1 |
RP11-346D14.1 |
| chr11_-_89540388 | 0.02 |
ENST00000532501.2 |
TRIM49 |
tripartite motif containing 49 |
| chr9_-_72287191 | 0.01 |
ENST00000265381.4 |
APBA1 |
amyloid beta (A4) precursor protein-binding, family A, member 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.7 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.1 | 0.3 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 0.3 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.3 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.1 | GO:0038188 | cholecystokinin signaling pathway(GO:0038188) |
| 0.0 | 0.4 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 0.1 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.2 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 0.2 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.9 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.2 | GO:0071420 | cellular response to histamine(GO:0071420) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.8 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 0.3 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.4 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0005602 | complement component C1 complex(GO:0005602) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.8 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.3 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.0 | 0.1 | GO:0004951 | cholecystokinin receptor activity(GO:0004951) |
| 0.0 | 0.1 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.0 | 0.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.2 | GO:0030884 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 0.2 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.2 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |