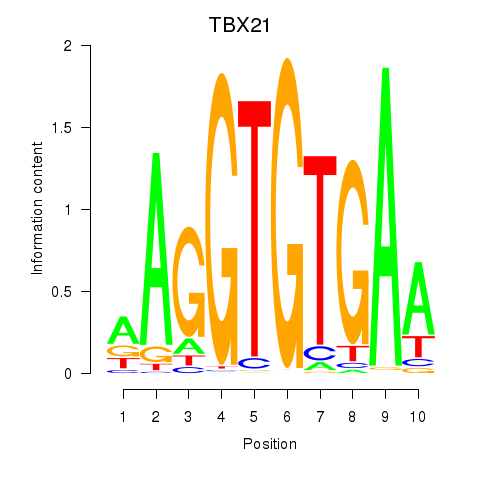
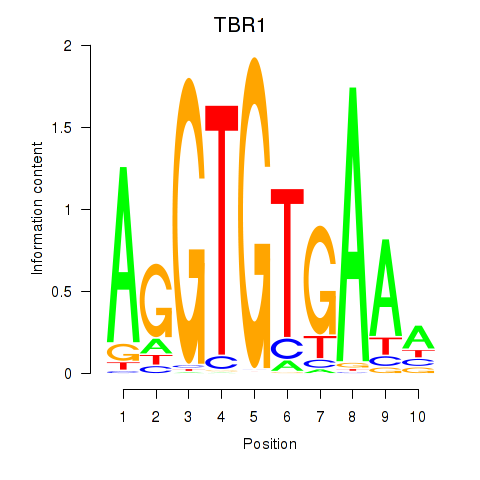
Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for TBX21_TBR1
Z-value: 0.90
Transcription factors associated with TBX21_TBR1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX21
|
ENSG00000073861.2 | TBX21 |
|
TBR1
|
ENSG00000136535.10 | TBR1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TBX21 | hg19_v2_chr17_+_45810594_45810610 | 0.87 | 4.6e-03 | Click! |
| TBR1 | hg19_v2_chr2_+_162272605_162272753 | -0.63 | 9.8e-02 | Click! |
Activity profile of TBX21_TBR1 motif
Sorted Z-values of TBX21_TBR1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TBX21_TBR1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_12862604 | 2.18 |
ENST00000553030.1 |
BEST2 |
bestrophin 2 |
| chr4_+_124317940 | 1.69 |
ENST00000505319.1 ENST00000339241.1 |
SPRY1 |
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
| chr3_-_100712352 | 1.59 |
ENST00000471714.1 ENST00000284322.5 |
ABI3BP |
ABI family, member 3 (NESH) binding protein |
| chr19_+_12862486 | 1.39 |
ENST00000549706.1 |
BEST2 |
bestrophin 2 |
| chr1_-_85930246 | 1.36 |
ENST00000426972.3 |
DDAH1 |
dimethylarginine dimethylaminohydrolase 1 |
| chr3_-_66551351 | 1.12 |
ENST00000273261.3 |
LRIG1 |
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr4_-_89619386 | 1.10 |
ENST00000323061.5 |
NAP1L5 |
nucleosome assembly protein 1-like 5 |
| chr3_-_66551397 | 1.09 |
ENST00000383703.3 |
LRIG1 |
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr2_-_175870085 | 1.06 |
ENST00000409156.3 |
CHN1 |
chimerin 1 |
| chr2_-_175869936 | 1.05 |
ENST00000409900.3 |
CHN1 |
chimerin 1 |
| chr12_-_91576561 | 0.99 |
ENST00000547568.2 ENST00000552962.1 |
DCN |
decorin |
| chr9_+_17134980 | 0.95 |
ENST00000380647.3 |
CNTLN |
centlein, centrosomal protein |
| chr12_-_91576429 | 0.92 |
ENST00000552145.1 ENST00000546745.1 |
DCN |
decorin |
| chr2_-_200322723 | 0.86 |
ENST00000417098.1 |
SATB2 |
SATB homeobox 2 |
| chr10_+_31608054 | 0.80 |
ENST00000320985.10 ENST00000361642.5 ENST00000560721.2 ENST00000558440.1 ENST00000424869.1 ENST00000542815.3 |
ZEB1 |
zinc finger E-box binding homeobox 1 |
| chr1_+_38022513 | 0.78 |
ENST00000296218.7 |
DNALI1 |
dynein, axonemal, light intermediate chain 1 |
| chr6_-_46459675 | 0.73 |
ENST00000306764.7 |
RCAN2 |
regulator of calcineurin 2 |
| chr12_+_65672702 | 0.72 |
ENST00000538045.1 ENST00000535239.1 |
MSRB3 |
methionine sulfoxide reductase B3 |
| chr1_-_163172625 | 0.69 |
ENST00000527988.1 ENST00000531476.1 ENST00000530507.1 |
RGS5 |
regulator of G-protein signaling 5 |
| chr5_+_140571902 | 0.68 |
ENST00000239446.4 |
PCDHB10 |
protocadherin beta 10 |
| chr14_+_95078714 | 0.68 |
ENST00000393078.3 ENST00000393080.4 ENST00000467132.1 |
SERPINA3 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 |
| chr22_+_31003190 | 0.67 |
ENST00000407817.3 |
TCN2 |
transcobalamin II |
| chr22_+_31003133 | 0.67 |
ENST00000405742.3 |
TCN2 |
transcobalamin II |
| chr16_+_53133070 | 0.66 |
ENST00000565832.1 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
| chr9_+_90112117 | 0.61 |
ENST00000358077.5 |
DAPK1 |
death-associated protein kinase 1 |
| chr13_+_76378305 | 0.60 |
ENST00000526371.1 ENST00000526528.1 |
LMO7 |
LIM domain 7 |
| chr15_+_41913690 | 0.59 |
ENST00000563576.1 |
MGA |
MGA, MAX dimerization protein |
| chr11_-_82782861 | 0.57 |
ENST00000524635.1 ENST00000526205.1 ENST00000527633.1 ENST00000533486.1 ENST00000533276.2 |
RAB30 |
RAB30, member RAS oncogene family |
| chr12_-_90024360 | 0.54 |
ENST00000393164.2 |
ATP2B1 |
ATPase, Ca++ transporting, plasma membrane 1 |
| chr3_-_120170052 | 0.54 |
ENST00000295633.3 |
FSTL1 |
follistatin-like 1 |
| chr5_-_147286065 | 0.53 |
ENST00000318315.4 ENST00000515291.1 |
C5orf46 |
chromosome 5 open reading frame 46 |
| chr1_+_38022572 | 0.52 |
ENST00000541606.1 |
DNALI1 |
dynein, axonemal, light intermediate chain 1 |
| chr3_-_179169330 | 0.51 |
ENST00000232564.3 |
GNB4 |
guanine nucleotide binding protein (G protein), beta polypeptide 4 |
| chr13_+_76378357 | 0.50 |
ENST00000489941.2 ENST00000525373.1 |
LMO7 |
LIM domain 7 |
| chr22_+_35776828 | 0.50 |
ENST00000216117.8 |
HMOX1 |
heme oxygenase (decycling) 1 |
| chr2_-_79313973 | 0.48 |
ENST00000454188.1 |
REG1B |
regenerating islet-derived 1 beta |
| chr12_-_91348949 | 0.47 |
ENST00000358859.2 |
CCER1 |
coiled-coil glutamate-rich protein 1 |
| chr9_+_134378289 | 0.46 |
ENST00000423007.1 ENST00000404875.2 ENST00000441334.1 ENST00000341012.7 ENST00000372228.3 ENST00000402686.3 ENST00000419118.2 ENST00000541219.1 ENST00000354713.4 ENST00000418774.1 ENST00000415075.1 ENST00000448212.1 ENST00000430619.1 |
POMT1 |
protein-O-mannosyltransferase 1 |
| chr2_+_217498105 | 0.46 |
ENST00000233809.4 |
IGFBP2 |
insulin-like growth factor binding protein 2, 36kDa |
| chr12_+_65672423 | 0.45 |
ENST00000355192.3 ENST00000308259.5 ENST00000540804.1 ENST00000535664.1 ENST00000541189.1 |
MSRB3 |
methionine sulfoxide reductase B3 |
| chr14_+_21467414 | 0.44 |
ENST00000554422.1 ENST00000298681.4 |
SLC39A2 |
solute carrier family 39 (zinc transporter), member 2 |
| chr1_+_201708992 | 0.44 |
ENST00000367295.1 |
NAV1 |
neuron navigator 1 |
| chr6_-_153452356 | 0.44 |
ENST00000206262.1 |
RGS17 |
regulator of G-protein signaling 17 |
| chr15_-_83316087 | 0.43 |
ENST00000568757.1 |
CPEB1 |
cytoplasmic polyadenylation element binding protein 1 |
| chr11_+_61447845 | 0.43 |
ENST00000257215.5 |
DAGLA |
diacylglycerol lipase, alpha |
| chr1_-_46598284 | 0.43 |
ENST00000423209.1 ENST00000262741.5 |
PIK3R3 |
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr1_-_46598371 | 0.43 |
ENST00000372006.1 ENST00000425892.1 ENST00000420542.1 ENST00000354242.4 ENST00000340332.6 |
PIK3R3 |
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr12_-_96184533 | 0.43 |
ENST00000343702.4 ENST00000344911.4 |
NTN4 |
netrin 4 |
| chr17_-_8066843 | 0.42 |
ENST00000404970.3 |
VAMP2 |
vesicle-associated membrane protein 2 (synaptobrevin 2) |
| chr14_-_65569244 | 0.41 |
ENST00000557277.1 ENST00000556892.1 |
MAX |
MYC associated factor X |
| chr11_-_82782952 | 0.40 |
ENST00000534141.1 |
RAB30 |
RAB30, member RAS oncogene family |
| chr9_-_35815013 | 0.40 |
ENST00000259667.5 |
HINT2 |
histidine triad nucleotide binding protein 2 |
| chr1_+_26146397 | 0.39 |
ENST00000374303.2 ENST00000533762.1 ENST00000529116.1 ENST00000474295.1 ENST00000488327.2 ENST00000472643.1 ENST00000526894.1 ENST00000524618.1 ENST00000374307.5 |
MTFR1L |
mitochondrial fission regulator 1-like |
| chr6_+_132873832 | 0.39 |
ENST00000275200.1 |
TAAR8 |
trace amine associated receptor 8 |
| chr17_-_15165854 | 0.39 |
ENST00000395936.1 ENST00000395938.2 |
PMP22 |
peripheral myelin protein 22 |
| chr15_-_83316254 | 0.39 |
ENST00000567678.1 ENST00000450751.2 |
CPEB1 |
cytoplasmic polyadenylation element binding protein 1 |
| chr8_-_27457494 | 0.38 |
ENST00000521770.1 |
CLU |
clusterin |
| chr5_+_54320078 | 0.38 |
ENST00000231009.2 |
GZMK |
granzyme K (granzyme 3; tryptase II) |
| chr4_-_186732048 | 0.38 |
ENST00000448662.2 ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr15_-_90294523 | 0.38 |
ENST00000300057.4 |
MESP1 |
mesoderm posterior 1 homolog (mouse) |
| chr6_+_45390222 | 0.37 |
ENST00000359524.5 |
RUNX2 |
runt-related transcription factor 2 |
| chr1_+_26146674 | 0.37 |
ENST00000525713.1 ENST00000374301.3 |
MTFR1L |
mitochondrial fission regulator 1-like |
| chr12_+_57853918 | 0.36 |
ENST00000532291.1 ENST00000543426.1 ENST00000228682.2 ENST00000546141.1 |
GLI1 |
GLI family zinc finger 1 |
| chr2_-_157189180 | 0.36 |
ENST00000539077.1 ENST00000424077.1 ENST00000426264.1 ENST00000339562.4 ENST00000421709.1 |
NR4A2 |
nuclear receptor subfamily 4, group A, member 2 |
| chr12_-_56120838 | 0.35 |
ENST00000548160.1 |
CD63 |
CD63 molecule |
| chr15_+_44119159 | 0.35 |
ENST00000263795.6 ENST00000381246.2 ENST00000452115.1 |
WDR76 |
WD repeat domain 76 |
| chr6_-_154751629 | 0.35 |
ENST00000424998.1 |
CNKSR3 |
CNKSR family member 3 |
| chr11_+_65266507 | 0.34 |
ENST00000544868.1 |
MALAT1 |
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chr10_+_90660832 | 0.34 |
ENST00000371924.1 |
STAMBPL1 |
STAM binding protein-like 1 |
| chr6_+_45389893 | 0.33 |
ENST00000371432.3 |
RUNX2 |
runt-related transcription factor 2 |
| chr19_-_44405941 | 0.33 |
ENST00000587128.1 |
RP11-15A1.3 |
RP11-15A1.3 |
| chr4_+_160188889 | 0.33 |
ENST00000264431.4 |
RAPGEF2 |
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr8_-_93107443 | 0.32 |
ENST00000360348.2 ENST00000520428.1 ENST00000518992.1 ENST00000520556.1 ENST00000518317.1 ENST00000521319.1 ENST00000521375.1 ENST00000518449.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr5_-_68628543 | 0.32 |
ENST00000396496.2 ENST00000511257.1 ENST00000383374.2 |
CCDC125 |
coiled-coil domain containing 125 |
| chr17_-_76356148 | 0.32 |
ENST00000587578.1 ENST00000330871.2 |
SOCS3 |
suppressor of cytokine signaling 3 |
| chr15_+_60296421 | 0.32 |
ENST00000396057.4 |
FOXB1 |
forkhead box B1 |
| chr5_-_131132614 | 0.32 |
ENST00000307968.7 ENST00000307954.8 |
FNIP1 |
folliculin interacting protein 1 |
| chr19_-_36236292 | 0.30 |
ENST00000378975.3 ENST00000412391.2 ENST00000292879.5 |
U2AF1L4 |
U2 small nuclear RNA auxiliary factor 1-like 4 |
| chr6_+_39760129 | 0.30 |
ENST00000274867.4 |
DAAM2 |
dishevelled associated activator of morphogenesis 2 |
| chr2_-_152146385 | 0.30 |
ENST00000414946.1 ENST00000243346.5 |
NMI |
N-myc (and STAT) interactor |
| chr2_+_162087577 | 0.30 |
ENST00000439442.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr13_+_114238997 | 0.29 |
ENST00000538138.1 ENST00000375370.5 |
TFDP1 |
transcription factor Dp-1 |
| chr19_+_46367518 | 0.29 |
ENST00000302177.2 |
FOXA3 |
forkhead box A3 |
| chrX_+_51636629 | 0.29 |
ENST00000375722.1 ENST00000326587.7 ENST00000375695.2 |
MAGED1 |
melanoma antigen family D, 1 |
| chr1_+_144989309 | 0.29 |
ENST00000596396.1 |
AL590452.1 |
Uncharacterized protein |
| chr10_+_45406627 | 0.29 |
ENST00000389583.4 |
TMEM72 |
transmembrane protein 72 |
| chr1_+_145507587 | 0.28 |
ENST00000330165.8 ENST00000369307.3 |
RBM8A |
RNA binding motif protein 8A |
| chr9_+_124048864 | 0.28 |
ENST00000545652.1 |
GSN |
gelsolin |
| chr19_-_50529193 | 0.28 |
ENST00000596445.1 ENST00000599538.1 |
VRK3 |
vaccinia related kinase 3 |
| chr7_-_138386097 | 0.28 |
ENST00000421622.1 |
SVOPL |
SVOP-like |
| chr1_-_1009683 | 0.28 |
ENST00000453464.2 |
RNF223 |
ring finger protein 223 |
| chr1_-_28969517 | 0.27 |
ENST00000263974.4 ENST00000373824.4 |
TAF12 |
TAF12 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 20kDa |
| chr8_+_22428457 | 0.27 |
ENST00000517962.1 |
SORBS3 |
sorbin and SH3 domain containing 3 |
| chr22_-_31688381 | 0.26 |
ENST00000487265.2 |
PIK3IP1 |
phosphoinositide-3-kinase interacting protein 1 |
| chrX_-_140271249 | 0.26 |
ENST00000370526.2 |
LDOC1 |
leucine zipper, down-regulated in cancer 1 |
| chr14_-_45603657 | 0.25 |
ENST00000396062.3 |
FKBP3 |
FK506 binding protein 3, 25kDa |
| chr17_+_58755184 | 0.25 |
ENST00000589222.1 ENST00000407086.3 ENST00000390652.5 |
BCAS3 |
breast carcinoma amplified sequence 3 |
| chr3_-_49466686 | 0.25 |
ENST00000273598.3 ENST00000436744.2 |
NICN1 |
nicolin 1 |
| chr7_-_120497178 | 0.25 |
ENST00000441017.1 ENST00000424710.1 ENST00000433758.1 |
TSPAN12 |
tetraspanin 12 |
| chr2_+_201994042 | 0.25 |
ENST00000417748.1 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chr1_+_39456895 | 0.24 |
ENST00000432648.3 ENST00000446189.2 ENST00000372984.4 |
AKIRIN1 |
akirin 1 |
| chr16_+_12995468 | 0.24 |
ENST00000424107.3 ENST00000558583.1 ENST00000558318.1 |
SHISA9 |
shisa family member 9 |
| chr22_-_43042955 | 0.24 |
ENST00000402438.1 |
CYB5R3 |
cytochrome b5 reductase 3 |
| chr10_+_94451574 | 0.24 |
ENST00000492654.2 |
HHEX |
hematopoietically expressed homeobox |
| chr9_+_34990219 | 0.23 |
ENST00000541010.1 ENST00000454002.2 ENST00000545841.1 |
DNAJB5 |
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr4_+_26585686 | 0.23 |
ENST00000505206.1 ENST00000511789.1 |
TBC1D19 |
TBC1 domain family, member 19 |
| chr17_-_26662464 | 0.23 |
ENST00000579419.1 ENST00000585313.1 ENST00000395418.3 ENST00000578985.1 ENST00000577498.1 ENST00000585089.1 ENST00000357896.3 |
IFT20 |
intraflagellar transport 20 homolog (Chlamydomonas) |
| chr17_-_26662440 | 0.23 |
ENST00000578122.1 |
IFT20 |
intraflagellar transport 20 homolog (Chlamydomonas) |
| chr19_-_7990991 | 0.23 |
ENST00000318978.4 |
CTXN1 |
cortexin 1 |
| chr4_-_8073705 | 0.23 |
ENST00000514025.1 |
ABLIM2 |
actin binding LIM protein family, member 2 |
| chr4_+_166300084 | 0.22 |
ENST00000402744.4 |
CPE |
carboxypeptidase E |
| chr22_+_40441456 | 0.22 |
ENST00000402203.1 |
TNRC6B |
trinucleotide repeat containing 6B |
| chr8_-_93107696 | 0.22 |
ENST00000436581.2 ENST00000520583.1 ENST00000519061.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr6_-_84937314 | 0.22 |
ENST00000257766.4 ENST00000403245.3 |
KIAA1009 |
KIAA1009 |
| chr16_+_27413483 | 0.22 |
ENST00000337929.3 ENST00000564089.1 |
IL21R |
interleukin 21 receptor |
| chr2_-_3584430 | 0.22 |
ENST00000438482.1 ENST00000422961.1 |
AC108488.4 |
AC108488.4 |
| chr1_+_174843548 | 0.22 |
ENST00000478442.1 ENST00000465412.1 |
RABGAP1L |
RAB GTPase activating protein 1-like |
| chr11_+_73358594 | 0.21 |
ENST00000227214.6 ENST00000398494.4 ENST00000543085.1 |
PLEKHB1 |
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr2_+_219247021 | 0.21 |
ENST00000539932.1 |
SLC11A1 |
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 1 |
| chrX_+_102862834 | 0.21 |
ENST00000372627.5 ENST00000243286.3 |
TCEAL3 |
transcription elongation factor A (SII)-like 3 |
| chr6_+_131894284 | 0.21 |
ENST00000368087.3 ENST00000356962.2 |
ARG1 |
arginase 1 |
| chr9_+_34329493 | 0.21 |
ENST00000379158.2 ENST00000346365.4 ENST00000379155.5 |
NUDT2 |
nudix (nucleoside diphosphate linked moiety X)-type motif 2 |
| chrX_-_69509738 | 0.21 |
ENST00000374454.1 ENST00000239666.4 |
PDZD11 |
PDZ domain containing 11 |
| chr7_-_76255444 | 0.21 |
ENST00000454397.1 |
POMZP3 |
POM121 and ZP3 fusion |
| chr8_-_6914251 | 0.21 |
ENST00000330590.2 |
DEFA5 |
defensin, alpha 5, Paneth cell-specific |
| chr7_-_138363824 | 0.21 |
ENST00000419765.3 |
SVOPL |
SVOP-like |
| chr10_+_118305435 | 0.21 |
ENST00000369221.2 |
PNLIP |
pancreatic lipase |
| chr20_+_18125727 | 0.21 |
ENST00000489634.2 |
CSRP2BP |
CSRP2 binding protein |
| chr1_+_213123976 | 0.20 |
ENST00000366965.2 ENST00000366967.2 |
VASH2 |
vasohibin 2 |
| chr13_+_42031679 | 0.20 |
ENST00000379359.3 |
RGCC |
regulator of cell cycle |
| chr5_+_94727048 | 0.20 |
ENST00000283357.5 |
FAM81B |
family with sequence similarity 81, member B |
| chr17_+_9066252 | 0.20 |
ENST00000436734.1 |
NTN1 |
netrin 1 |
| chr3_-_146213722 | 0.20 |
ENST00000336685.2 ENST00000489015.1 |
PLSCR2 |
phospholipid scramblase 2 |
| chr9_+_125273081 | 0.20 |
ENST00000335302.5 |
OR1J2 |
olfactory receptor, family 1, subfamily J, member 2 |
| chr5_+_137419581 | 0.20 |
ENST00000506684.1 ENST00000504809.1 ENST00000398754.1 |
WNT8A |
wingless-type MMTV integration site family, member 8A |
| chr2_-_166060571 | 0.19 |
ENST00000360093.3 |
SCN3A |
sodium channel, voltage-gated, type III, alpha subunit |
| chr1_-_113247543 | 0.19 |
ENST00000414971.1 ENST00000534717.1 |
RHOC |
ras homolog family member C |
| chr17_-_18585131 | 0.19 |
ENST00000443457.1 ENST00000583002.1 |
ZNF286B |
zinc finger protein 286B |
| chr8_-_42623924 | 0.19 |
ENST00000276410.2 |
CHRNA6 |
cholinergic receptor, nicotinic, alpha 6 (neuronal) |
| chr9_+_17135016 | 0.19 |
ENST00000425824.1 ENST00000262360.5 ENST00000380641.4 |
CNTLN |
centlein, centrosomal protein |
| chr5_+_6448736 | 0.19 |
ENST00000399816.3 |
UBE2QL1 |
ubiquitin-conjugating enzyme E2Q family-like 1 |
| chr3_+_42977846 | 0.19 |
ENST00000383748.4 |
KRBOX1 |
KRAB box domain containing 1 |
| chr4_+_100495864 | 0.19 |
ENST00000265517.5 ENST00000422897.2 |
MTTP |
microsomal triglyceride transfer protein |
| chr8_+_38586068 | 0.19 |
ENST00000443286.2 ENST00000520340.1 ENST00000518415.1 |
TACC1 |
transforming, acidic coiled-coil containing protein 1 |
| chr1_+_156105878 | 0.19 |
ENST00000508500.1 |
LMNA |
lamin A/C |
| chr16_+_21716284 | 0.19 |
ENST00000388957.3 |
OTOA |
otoancorin |
| chr2_-_61697862 | 0.19 |
ENST00000398571.2 |
USP34 |
ubiquitin specific peptidase 34 |
| chr9_+_33750667 | 0.19 |
ENST00000457896.1 ENST00000342836.4 ENST00000429677.3 |
PRSS3 |
protease, serine, 3 |
| chr17_+_75084717 | 0.19 |
ENST00000561721.2 ENST00000589827.1 ENST00000392476.2 |
SEC14L1 |
SEC14-like 1 (S. cerevisiae) |
| chr12_+_12938541 | 0.18 |
ENST00000356591.4 |
APOLD1 |
apolipoprotein L domain containing 1 |
| chr9_+_33750515 | 0.18 |
ENST00000361005.5 |
PRSS3 |
protease, serine, 3 |
| chr2_+_204192942 | 0.18 |
ENST00000295851.5 ENST00000261017.5 |
ABI2 |
abl-interactor 2 |
| chr8_-_42623747 | 0.18 |
ENST00000534622.1 |
CHRNA6 |
cholinergic receptor, nicotinic, alpha 6 (neuronal) |
| chr2_-_166060552 | 0.18 |
ENST00000283254.7 ENST00000453007.1 |
SCN3A |
sodium channel, voltage-gated, type III, alpha subunit |
| chr7_-_14942283 | 0.17 |
ENST00000402815.1 |
DGKB |
diacylglycerol kinase, beta 90kDa |
| chr13_+_53602894 | 0.17 |
ENST00000219022.2 |
OLFM4 |
olfactomedin 4 |
| chr17_+_15603447 | 0.17 |
ENST00000395893.2 |
ZNF286A |
Homo sapiens zinc finger protein 286A (ZNF286A), transcript variant 6, mRNA. |
| chr6_+_160221293 | 0.17 |
ENST00000610273.1 ENST00000392167.3 |
PNLDC1 |
poly(A)-specific ribonuclease (PARN)-like domain containing 1 |
| chr12_-_110511424 | 0.17 |
ENST00000548191.1 |
C12orf76 |
chromosome 12 open reading frame 76 |
| chr17_-_1553346 | 0.17 |
ENST00000301336.6 |
RILP |
Rab interacting lysosomal protein |
| chr2_+_85646054 | 0.17 |
ENST00000389938.2 |
SH2D6 |
SH2 domain containing 6 |
| chr6_-_33679452 | 0.17 |
ENST00000374231.4 ENST00000607484.1 ENST00000374214.3 |
UQCC2 |
ubiquinol-cytochrome c reductase complex assembly factor 2 |
| chr22_-_31688431 | 0.17 |
ENST00000402249.3 ENST00000443175.1 ENST00000215912.5 ENST00000441972.1 |
PIK3IP1 |
phosphoinositide-3-kinase interacting protein 1 |
| chr11_+_45907177 | 0.17 |
ENST00000241014.2 |
MAPK8IP1 |
mitogen-activated protein kinase 8 interacting protein 1 |
| chr8_+_38585704 | 0.17 |
ENST00000519416.1 ENST00000520615.1 |
TACC1 |
transforming, acidic coiled-coil containing protein 1 |
| chr12_-_76879852 | 0.17 |
ENST00000548341.1 |
OSBPL8 |
oxysterol binding protein-like 8 |
| chr11_-_64052111 | 0.16 |
ENST00000394532.3 ENST00000394531.3 ENST00000309032.3 |
BAD |
BCL2-associated agonist of cell death |
| chr14_+_77582905 | 0.16 |
ENST00000557408.1 |
TMEM63C |
transmembrane protein 63C |
| chr3_+_38035610 | 0.16 |
ENST00000465644.1 |
VILL |
villin-like |
| chr22_-_43042968 | 0.16 |
ENST00000407623.3 ENST00000396303.3 ENST00000438270.1 |
CYB5R3 |
cytochrome b5 reductase 3 |
| chr9_-_132805430 | 0.16 |
ENST00000446176.2 ENST00000355681.3 ENST00000420781.1 |
FNBP1 |
formin binding protein 1 |
| chr16_+_69221028 | 0.16 |
ENST00000336278.4 |
SNTB2 |
syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2) |
| chr9_+_35829208 | 0.15 |
ENST00000439587.2 ENST00000377991.4 |
TMEM8B |
transmembrane protein 8B |
| chr10_-_23003460 | 0.15 |
ENST00000376573.4 |
PIP4K2A |
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
| chrX_+_77166172 | 0.15 |
ENST00000343533.5 ENST00000350425.4 ENST00000341514.6 |
ATP7A |
ATPase, Cu++ transporting, alpha polypeptide |
| chr5_-_176433350 | 0.15 |
ENST00000377227.4 ENST00000377219.2 |
UIMC1 |
ubiquitin interaction motif containing 1 |
| chr13_+_46276441 | 0.15 |
ENST00000310521.1 ENST00000533564.1 |
SPERT |
spermatid associated |
| chr12_-_56120865 | 0.15 |
ENST00000548898.1 ENST00000552067.1 |
CD63 |
CD63 molecule |
| chr6_+_168418553 | 0.15 |
ENST00000354419.2 ENST00000351261.3 |
KIF25 |
kinesin family member 25 |
| chr11_+_18477369 | 0.15 |
ENST00000396213.3 ENST00000280706.2 |
LDHAL6A |
lactate dehydrogenase A-like 6A |
| chr17_-_56609302 | 0.15 |
ENST00000581607.1 ENST00000317256.6 ENST00000426861.1 ENST00000580809.1 ENST00000577729.1 ENST00000583291.1 |
SEPT4 |
septin 4 |
| chr1_-_150780757 | 0.15 |
ENST00000271651.3 |
CTSK |
cathepsin K |
| chr11_-_114271139 | 0.15 |
ENST00000325636.4 |
C11orf71 |
chromosome 11 open reading frame 71 |
| chr8_-_72268889 | 0.15 |
ENST00000388742.4 |
EYA1 |
eyes absent homolog 1 (Drosophila) |
| chr11_-_236326 | 0.15 |
ENST00000525237.1 ENST00000532956.1 ENST00000525319.1 ENST00000524564.1 ENST00000382743.4 |
SIRT3 |
sirtuin 3 |
| chr11_-_6440624 | 0.14 |
ENST00000311051.3 |
APBB1 |
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chrX_-_19689106 | 0.14 |
ENST00000379716.1 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
| chr3_-_100712292 | 0.14 |
ENST00000495063.1 ENST00000530539.1 |
ABI3BP |
ABI family, member 3 (NESH) binding protein |
| chr11_+_122709200 | 0.14 |
ENST00000227348.4 |
CRTAM |
cytotoxic and regulatory T cell molecule |
| chr11_+_66278080 | 0.14 |
ENST00000318312.7 ENST00000526815.1 ENST00000537537.1 ENST00000525809.1 ENST00000455748.2 ENST00000393994.2 |
BBS1 |
Bardet-Biedl syndrome 1 |
| chr6_-_30585009 | 0.14 |
ENST00000376511.2 |
PPP1R10 |
protein phosphatase 1, regulatory subunit 10 |
| chr5_-_131132658 | 0.14 |
ENST00000514667.1 ENST00000511848.1 ENST00000510461.1 |
CTC-432M15.3 FNIP1 |
Folliculin-interacting protein 1 folliculin interacting protein 1 |
| chrX_+_102631844 | 0.14 |
ENST00000372634.1 ENST00000299872.7 |
NGFRAP1 |
nerve growth factor receptor (TNFRSF16) associated protein 1 |
| chr11_+_55653396 | 0.14 |
ENST00000244891.3 |
TRIM51 |
tripartite motif-containing 51 |
| chr16_-_30546141 | 0.13 |
ENST00000535210.1 ENST00000395094.3 |
ZNF747 |
zinc finger protein 747 |
| chr20_+_36661910 | 0.13 |
ENST00000373433.4 |
RPRD1B |
regulation of nuclear pre-mRNA domain containing 1B |
| chr1_+_89829610 | 0.13 |
ENST00000370456.4 ENST00000535065.1 |
GBP6 |
guanylate binding protein family, member 6 |
| chr7_-_99766191 | 0.13 |
ENST00000423751.1 ENST00000360039.4 |
GAL3ST4 |
galactose-3-O-sulfotransferase 4 |
| chr1_-_241799232 | 0.13 |
ENST00000366553.1 |
CHML |
choroideremia-like (Rab escort protein 2) |
| chr3_+_14058794 | 0.13 |
ENST00000424053.1 ENST00000528067.1 ENST00000429201.1 |
TPRXL |
tetra-peptide repeat homeobox-like |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.4 | 2.2 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.2 | 0.4 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.2 | 0.5 | GO:0006788 | heme oxidation(GO:0006788) smooth muscle hyperplasia(GO:0014806) negative regulation of mast cell cytokine production(GO:0032764) |
| 0.1 | 1.9 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 1.3 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.1 | 0.4 | GO:0060913 | cardiac cell fate determination(GO:0060913) negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.4 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.1 | 0.2 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.1 | 0.5 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.1 | 0.9 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.1 | 1.7 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.1 | 0.3 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.1 | 0.4 | GO:0098917 | retrograde trans-synaptic signaling(GO:0098917) |
| 0.1 | 1.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.3 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 0.5 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.1 | 0.3 | GO:0055073 | cadmium ion homeostasis(GO:0055073) divalent metal ion export(GO:0070839) |
| 0.1 | 0.9 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 0.4 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.1 | 0.3 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 0.4 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.1 | 0.4 | GO:0061317 | canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) |
| 0.1 | 0.3 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.1 | 0.4 | GO:0021938 | ventral midline development(GO:0007418) smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.1 | 0.1 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.1 | 0.7 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.5 | GO:1901660 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export(GO:1901660) calcium ion export from cell(GO:1990034) |
| 0.1 | 0.3 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.1 | 0.4 | GO:0061517 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.1 | 1.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 0.2 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.1 | 0.1 | GO:0002519 | natural killer cell tolerance induction(GO:0002519) |
| 0.1 | 0.8 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.2 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.1 | 0.3 | GO:0044855 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.1 | 0.2 | GO:0030070 | insulin processing(GO:0030070) |
| 0.1 | 0.7 | GO:2000680 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.1 | 0.2 | GO:0052151 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.1 | 0.4 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.1 | 0.2 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.1 | 0.2 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.0 | 0.3 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 2.1 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.1 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.1 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.5 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.1 | GO:0002325 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.0 | 0.1 | GO:0060921 | sinoatrial node development(GO:0003163) sinoatrial node cell differentiation(GO:0060921) |
| 0.0 | 0.2 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.2 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.2 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.4 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.1 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.0 | 0.1 | GO:2000777 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.0 | 0.2 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 0.1 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.0 | 0.2 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.0 | 1.0 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 | 0.2 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.7 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.1 | GO:0042704 | detection of oxygen(GO:0003032) uterine wall breakdown(GO:0042704) |
| 0.0 | 0.1 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 0.0 | 0.3 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.1 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.0 | 0.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.1 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 0.0 | 0.1 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.2 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:0003068 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.6 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.2 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.0 | 0.1 | GO:0009447 | putrescine catabolic process(GO:0009447) |
| 0.0 | 3.6 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.1 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.0 | 0.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.3 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.1 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.1 | GO:0072233 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.1 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.0 | 0.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.1 | GO:0072695 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.0 | 0.1 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.0 | 0.7 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.4 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.4 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.2 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.1 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.2 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.4 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.1 | GO:0002860 | positive regulation of response to tumor cell(GO:0002836) positive regulation of immune response to tumor cell(GO:0002839) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.1 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.1 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.0 | 0.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.2 | GO:0098856 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.0 | 0.0 | GO:0043449 | olfactory learning(GO:0008355) cellular alkene metabolic process(GO:0043449) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0002254 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 0.0 | 0.1 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.0 | 0.0 | GO:0033212 | iron assimilation(GO:0033212) iron assimilation by chelation and transport(GO:0033214) positive regulation of bone mineralization involved in bone maturation(GO:1900159) negative regulation of tumor necrosis factor (ligand) superfamily member 11 production(GO:2000308) |
| 0.0 | 0.1 | GO:0009407 | toxin catabolic process(GO:0009407) mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) secondary metabolite catabolic process(GO:0090487) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.0 | 0.2 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.1 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 | 0.1 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.0 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.0 | 0.8 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.0 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.0 | 0.3 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.1 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 0.0 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.0 | 0.1 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.3 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.3 | GO:1900120 | regulation of receptor binding(GO:1900120) |
| 0.0 | 0.3 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.5 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.2 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.3 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.3 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.2 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 0.4 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.3 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.5 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.2 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.6 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 1.9 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.4 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 0.7 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 1.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.4 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.3 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.2 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 3.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.8 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.9 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.8 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.5 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.4 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.3 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.2 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.0 | 0.1 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 0.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 1.0 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.2 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.1 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.0 | 0.2 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 1.3 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.4 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) ripoptosome(GO:0097342) |
| 0.0 | 0.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.0 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.5 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.9 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.5 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.1 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 0.1 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.2 | 1.4 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.2 | 0.5 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.4 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 0.7 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 1.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.5 | GO:0002046 | opsin binding(GO:0002046) |
| 0.1 | 0.3 | GO:0051139 | metal ion:proton antiporter activity(GO:0051139) |
| 0.1 | 0.3 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.1 | 0.5 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 0.5 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 0.4 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.3 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.1 | 0.7 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 1.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 0.2 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 0.2 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.3 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.4 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.5 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.8 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.0 | 0.3 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.1 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.0 | 0.1 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 3.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.0 | 0.4 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 1.0 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.0 | 1.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 3.6 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.7 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.7 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.4 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.3 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.2 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.3 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.9 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) acetylcholine binding(GO:0042166) |
| 0.0 | 0.2 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.0 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) (R)-2-hydroxyglutarate dehydrogenase activity(GO:0051990) |
| 0.0 | 0.4 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.2 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.5 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.0 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0016594 | glycine binding(GO:0016594) |
| 0.0 | 0.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.0 | 0.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.4 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 3.0 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.2 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.8 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.0 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.5 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.7 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.8 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 0.9 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 1.8 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 1.0 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.7 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.4 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.7 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.7 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 2.2 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.4 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.4 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.1 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.0 | 0.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |