Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for TCF7
Z-value: 0.56
Transcription factors associated with TCF7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TCF7
|
ENSG00000081059.15 | TCF7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TCF7 | hg19_v2_chr5_+_133477905_133477956 | -0.68 | 6.3e-02 | Click! |
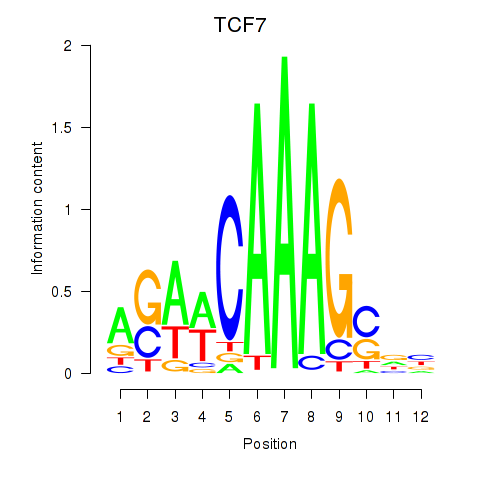
Activity profile of TCF7 motif
Sorted Z-values of TCF7 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TCF7
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_108422926 | 0.28 |
ENST00000428840.1 ENST00000526312.1 |
EXPH5 |
exophilin 5 |
| chr1_-_209824643 | 0.27 |
ENST00000391911.1 ENST00000415782.1 |
LAMB3 |
laminin, beta 3 |
| chr16_+_68678739 | 0.27 |
ENST00000264012.4 |
CDH3 |
cadherin 3, type 1, P-cadherin (placental) |
| chr19_+_45281118 | 0.25 |
ENST00000270279.3 ENST00000341505.4 |
CBLC |
Cbl proto-oncogene C, E3 ubiquitin protein ligase |
| chr5_+_140165876 | 0.24 |
ENST00000504120.2 ENST00000394633.3 ENST00000378133.3 |
PCDHA1 |
protocadherin alpha 1 |
| chr1_+_156254070 | 0.24 |
ENST00000405535.2 ENST00000456810.1 |
TMEM79 |
transmembrane protein 79 |
| chr17_+_48610074 | 0.23 |
ENST00000503690.1 ENST00000514874.1 ENST00000537145.1 ENST00000541226.1 |
EPN3 |
epsin 3 |
| chr5_-_16936340 | 0.22 |
ENST00000507288.1 ENST00000513610.1 |
MYO10 |
myosin X |
| chr1_-_205649580 | 0.21 |
ENST00000367145.3 |
SLC45A3 |
solute carrier family 45, member 3 |
| chr8_-_42234745 | 0.20 |
ENST00000220812.2 |
DKK4 |
dickkopf WNT signaling pathway inhibitor 4 |
| chr17_-_39580775 | 0.20 |
ENST00000225550.3 |
KRT37 |
keratin 37 |
| chr3_-_190167571 | 0.20 |
ENST00000354905.2 |
TMEM207 |
transmembrane protein 207 |
| chr6_+_125540951 | 0.18 |
ENST00000524679.1 |
TPD52L1 |
tumor protein D52-like 1 |
| chr7_-_16840820 | 0.18 |
ENST00000450569.1 |
AGR2 |
anterior gradient 2 |
| chr11_-_87908600 | 0.18 |
ENST00000531138.1 ENST00000526372.1 ENST00000243662.6 |
RAB38 |
RAB38, member RAS oncogene family |
| chr6_-_11779174 | 0.18 |
ENST00000379413.2 |
ADTRP |
androgen-dependent TFPI-regulating protein |
| chr11_+_120039685 | 0.17 |
ENST00000530303.1 ENST00000319763.1 |
AP000679.2 |
Uncharacterized protein |
| chr3_+_36421826 | 0.16 |
ENST00000273183.3 |
STAC |
SH3 and cysteine rich domain |
| chr1_-_153013588 | 0.16 |
ENST00000360379.3 |
SPRR2D |
small proline-rich protein 2D |
| chr1_-_153029980 | 0.16 |
ENST00000392653.2 |
SPRR2A |
small proline-rich protein 2A |
| chr3_+_111717600 | 0.16 |
ENST00000273368.4 |
TAGLN3 |
transgelin 3 |
| chr3_+_111718036 | 0.16 |
ENST00000455401.2 |
TAGLN3 |
transgelin 3 |
| chr1_+_15479054 | 0.16 |
ENST00000376014.3 ENST00000451326.2 |
TMEM51 |
transmembrane protein 51 |
| chr4_-_139163491 | 0.16 |
ENST00000280612.5 |
SLC7A11 |
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
| chr17_+_30813576 | 0.15 |
ENST00000313401.3 |
CDK5R1 |
cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
| chr10_-_116444371 | 0.15 |
ENST00000533213.2 ENST00000369252.4 |
ABLIM1 |
actin binding LIM protein 1 |
| chr1_+_15479021 | 0.15 |
ENST00000428417.1 |
TMEM51 |
transmembrane protein 51 |
| chr3_+_111718173 | 0.15 |
ENST00000494932.1 |
TAGLN3 |
transgelin 3 |
| chr6_-_11807277 | 0.15 |
ENST00000379415.2 |
ADTRP |
androgen-dependent TFPI-regulating protein |
| chr6_-_29527702 | 0.14 |
ENST00000377050.4 |
UBD |
ubiquitin D |
| chr3_+_111717511 | 0.14 |
ENST00000478951.1 ENST00000393917.2 |
TAGLN3 |
transgelin 3 |
| chr2_-_238499303 | 0.14 |
ENST00000409576.1 |
RAB17 |
RAB17, member RAS oncogene family |
| chr3_+_14989186 | 0.14 |
ENST00000435454.1 ENST00000323373.6 |
NR2C2 |
nuclear receptor subfamily 2, group C, member 2 |
| chr6_+_138188551 | 0.13 |
ENST00000237289.4 ENST00000433680.1 |
TNFAIP3 |
tumor necrosis factor, alpha-induced protein 3 |
| chr11_+_45943169 | 0.13 |
ENST00000529052.1 ENST00000531526.1 |
GYLTL1B |
glycosyltransferase-like 1B |
| chrX_+_135618258 | 0.13 |
ENST00000440515.1 ENST00000456412.1 |
VGLL1 |
vestigial like 1 (Drosophila) |
| chr8_+_56792377 | 0.13 |
ENST00000520220.2 |
LYN |
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog |
| chr2_+_26568965 | 0.13 |
ENST00000260585.7 ENST00000447170.1 |
EPT1 |
ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific) |
| chr6_+_13925318 | 0.12 |
ENST00000423553.2 ENST00000537388.1 |
RNF182 |
ring finger protein 182 |
| chr22_+_24105394 | 0.12 |
ENST00000305199.5 ENST00000382821.3 |
C22orf15 |
chromosome 22 open reading frame 15 |
| chr14_-_23624511 | 0.12 |
ENST00000529705.2 |
SLC7A8 |
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chr8_-_139926236 | 0.12 |
ENST00000303045.6 ENST00000435777.1 |
COL22A1 |
collagen, type XXII, alpha 1 |
| chr8_+_56792355 | 0.12 |
ENST00000519728.1 |
LYN |
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog |
| chr7_-_19184929 | 0.12 |
ENST00000275461.3 |
FERD3L |
Fer3-like bHLH transcription factor |
| chrX_-_117119243 | 0.11 |
ENST00000539496.1 ENST00000469946.1 |
KLHL13 |
kelch-like family member 13 |
| chr1_+_955448 | 0.11 |
ENST00000379370.2 |
AGRN |
agrin |
| chr2_-_238499337 | 0.11 |
ENST00000411462.1 ENST00000409822.1 |
RAB17 |
RAB17, member RAS oncogene family |
| chr12_+_10163231 | 0.11 |
ENST00000396502.1 ENST00000338896.5 |
CLEC12B |
C-type lectin domain family 12, member B |
| chr8_-_20161466 | 0.11 |
ENST00000381569.1 |
LZTS1 |
leucine zipper, putative tumor suppressor 1 |
| chr6_+_111408698 | 0.11 |
ENST00000368851.5 |
SLC16A10 |
solute carrier family 16 (aromatic amino acid transporter), member 10 |
| chr10_+_6622345 | 0.11 |
ENST00000445427.1 ENST00000455810.1 |
PRKCQ-AS1 |
PRKCQ antisense RNA 1 |
| chr5_-_82969405 | 0.11 |
ENST00000510978.1 |
HAPLN1 |
hyaluronan and proteoglycan link protein 1 |
| chr6_-_112408661 | 0.11 |
ENST00000368662.5 |
TUBE1 |
tubulin, epsilon 1 |
| chr17_-_56565736 | 0.10 |
ENST00000323777.3 |
HSF5 |
heat shock transcription factor family member 5 |
| chr4_-_74486347 | 0.10 |
ENST00000342081.3 |
RASSF6 |
Ras association (RalGDS/AF-6) domain family member 6 |
| chr14_-_85996332 | 0.10 |
ENST00000380722.1 |
RP11-497E19.1 |
RP11-497E19.1 |
| chr21_+_39628852 | 0.10 |
ENST00000398938.2 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr4_-_85419603 | 0.10 |
ENST00000295886.4 |
NKX6-1 |
NK6 homeobox 1 |
| chr15_-_52043722 | 0.10 |
ENST00000454181.2 |
LYSMD2 |
LysM, putative peptidoglycan-binding, domain containing 2 |
| chr15_-_89764929 | 0.10 |
ENST00000268125.5 |
RLBP1 |
retinaldehyde binding protein 1 |
| chr1_-_153113927 | 0.09 |
ENST00000368752.4 |
SPRR2B |
small proline-rich protein 2B |
| chr3_+_57875711 | 0.09 |
ENST00000442599.2 |
SLMAP |
sarcolemma associated protein |
| chr18_+_59992514 | 0.09 |
ENST00000269485.7 |
TNFRSF11A |
tumor necrosis factor receptor superfamily, member 11a, NFKB activator |
| chr12_+_6309963 | 0.09 |
ENST00000382515.2 |
CD9 |
CD9 molecule |
| chr6_+_13925098 | 0.09 |
ENST00000488300.1 ENST00000544682.1 ENST00000420478.2 |
RNF182 |
ring finger protein 182 |
| chr1_-_53793584 | 0.09 |
ENST00000354412.3 ENST00000347547.2 ENST00000306052.6 |
LRP8 |
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr18_+_59992527 | 0.09 |
ENST00000586569.1 |
TNFRSF11A |
tumor necrosis factor receptor superfamily, member 11a, NFKB activator |
| chr10_-_104211294 | 0.09 |
ENST00000239125.1 |
C10orf95 |
chromosome 10 open reading frame 95 |
| chr15_-_44486632 | 0.09 |
ENST00000484674.1 |
FRMD5 |
FERM domain containing 5 |
| chr5_+_34757309 | 0.09 |
ENST00000397449.1 |
RAI14 |
retinoic acid induced 14 |
| chr4_-_7069760 | 0.09 |
ENST00000264954.4 |
GRPEL1 |
GrpE-like 1, mitochondrial (E. coli) |
| chr3_+_118892362 | 0.09 |
ENST00000497685.1 ENST00000264234.3 |
UPK1B |
uroplakin 1B |
| chr10_-_61513201 | 0.08 |
ENST00000414264.1 ENST00000594536.1 |
LINC00948 |
long intergenic non-protein coding RNA 948 |
| chr7_-_36406750 | 0.08 |
ENST00000453212.1 ENST00000415803.2 ENST00000440378.1 ENST00000431396.1 ENST00000317020.6 ENST00000436884.1 |
KIAA0895 |
KIAA0895 |
| chr8_-_17579726 | 0.08 |
ENST00000381861.3 |
MTUS1 |
microtubule associated tumor suppressor 1 |
| chr7_-_16505440 | 0.08 |
ENST00000307068.4 |
SOSTDC1 |
sclerostin domain containing 1 |
| chr19_+_50084561 | 0.08 |
ENST00000246794.5 |
PRRG2 |
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr15_-_44487408 | 0.08 |
ENST00000402883.1 ENST00000417257.1 |
FRMD5 |
FERM domain containing 5 |
| chr4_-_100484825 | 0.08 |
ENST00000273962.3 ENST00000514547.1 ENST00000455368.2 |
TRMT10A |
tRNA methyltransferase 10 homolog A (S. cerevisiae) |
| chr9_+_103189405 | 0.08 |
ENST00000395067.2 |
MSANTD3 |
Myb/SANT-like DNA-binding domain containing 3 |
| chr12_-_71003568 | 0.08 |
ENST00000547715.1 ENST00000451516.2 ENST00000538708.1 ENST00000550857.1 ENST00000261266.5 |
PTPRB |
protein tyrosine phosphatase, receptor type, B |
| chr2_+_159651821 | 0.08 |
ENST00000309950.3 ENST00000409042.1 |
DAPL1 |
death associated protein-like 1 |
| chr8_+_27629459 | 0.08 |
ENST00000523566.1 |
ESCO2 |
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr1_-_62785054 | 0.08 |
ENST00000371153.4 |
KANK4 |
KN motif and ankyrin repeat domains 4 |
| chr12_+_32655110 | 0.08 |
ENST00000546442.1 ENST00000583694.1 |
FGD4 |
FYVE, RhoGEF and PH domain containing 4 |
| chr3_+_14989076 | 0.07 |
ENST00000413118.1 ENST00000425241.1 |
NR2C2 |
nuclear receptor subfamily 2, group C, member 2 |
| chr12_+_69080734 | 0.07 |
ENST00000378905.2 |
NUP107 |
nucleoporin 107kDa |
| chr12_-_8043736 | 0.07 |
ENST00000539924.1 |
SLC2A14 |
solute carrier family 2 (facilitated glucose transporter), member 14 |
| chr2_-_160761179 | 0.07 |
ENST00000554112.1 ENST00000553424.1 ENST00000263636.4 ENST00000504764.1 ENST00000505052.1 |
LY75 LY75-CD302 |
lymphocyte antigen 75 LY75-CD302 readthrough |
| chr4_+_56815102 | 0.07 |
ENST00000257287.4 |
CEP135 |
centrosomal protein 135kDa |
| chr7_+_139528952 | 0.07 |
ENST00000416849.2 ENST00000436047.2 ENST00000414508.2 ENST00000448866.1 |
TBXAS1 |
thromboxane A synthase 1 (platelet) |
| chr17_+_74372662 | 0.07 |
ENST00000591651.1 ENST00000545180.1 |
SPHK1 |
sphingosine kinase 1 |
| chr14_+_22520762 | 0.07 |
ENST00000390449.3 |
TRAV21 |
T cell receptor alpha variable 21 |
| chr22_+_29279552 | 0.07 |
ENST00000544604.2 |
ZNRF3 |
zinc and ring finger 3 |
| chr2_-_74780176 | 0.07 |
ENST00000409549.1 |
LOXL3 |
lysyl oxidase-like 3 |
| chr6_-_25874440 | 0.07 |
ENST00000361703.6 ENST00000397060.4 |
SLC17A3 |
solute carrier family 17 (organic anion transporter), member 3 |
| chr12_-_48963829 | 0.07 |
ENST00000301046.2 ENST00000549817.1 |
LALBA |
lactalbumin, alpha- |
| chr20_-_13971255 | 0.07 |
ENST00000284951.5 ENST00000378072.5 |
SEL1L2 |
sel-1 suppressor of lin-12-like 2 (C. elegans) |
| chr19_+_41949054 | 0.06 |
ENST00000378187.2 |
C19orf69 |
chromosome 19 open reading frame 69 |
| chr13_+_45694583 | 0.06 |
ENST00000340473.6 |
GTF2F2 |
general transcription factor IIF, polypeptide 2, 30kDa |
| chr14_-_35344093 | 0.06 |
ENST00000382422.2 |
BAZ1A |
bromodomain adjacent to zinc finger domain, 1A |
| chr5_+_140235469 | 0.06 |
ENST00000506939.2 ENST00000307360.5 |
PCDHA10 |
protocadherin alpha 10 |
| chr22_-_37403839 | 0.06 |
ENST00000402860.3 ENST00000381821.1 |
TEX33 |
testis expressed 33 |
| chr22_-_37403858 | 0.06 |
ENST00000405091.2 |
TEX33 |
testis expressed 33 |
| chr7_-_148725733 | 0.06 |
ENST00000286091.4 |
PDIA4 |
protein disulfide isomerase family A, member 4 |
| chr2_+_108994466 | 0.06 |
ENST00000272452.2 |
SULT1C4 |
sulfotransferase family, cytosolic, 1C, member 4 |
| chr12_-_10151773 | 0.06 |
ENST00000298527.6 ENST00000348658.4 |
CLEC1B |
C-type lectin domain family 1, member B |
| chr4_-_23735183 | 0.06 |
ENST00000507666.1 |
RP11-380P13.2 |
RP11-380P13.2 |
| chr5_+_140207536 | 0.06 |
ENST00000529310.1 ENST00000527624.1 |
PCDHA6 |
protocadherin alpha 6 |
| chr10_+_96698406 | 0.06 |
ENST00000260682.6 |
CYP2C9 |
cytochrome P450, family 2, subfamily C, polypeptide 9 |
| chr14_+_32798547 | 0.06 |
ENST00000557354.1 ENST00000557102.1 ENST00000557272.1 |
AKAP6 |
A kinase (PRKA) anchor protein 6 |
| chr21_-_32185570 | 0.06 |
ENST00000329621.4 |
KRTAP8-1 |
keratin associated protein 8-1 |
| chr19_+_10765003 | 0.06 |
ENST00000407004.3 ENST00000589998.1 ENST00000589600.1 |
ILF3 |
interleukin enhancer binding factor 3, 90kDa |
| chr1_-_155948318 | 0.06 |
ENST00000361247.4 |
ARHGEF2 |
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
| chr6_+_159084188 | 0.06 |
ENST00000367081.3 |
SYTL3 |
synaptotagmin-like 3 |
| chr22_-_37880543 | 0.06 |
ENST00000442496.1 |
MFNG |
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr2_+_24150180 | 0.06 |
ENST00000404924.1 |
UBXN2A |
UBX domain protein 2A |
| chr17_-_17485731 | 0.06 |
ENST00000395783.1 |
PEMT |
phosphatidylethanolamine N-methyltransferase |
| chr4_-_80329356 | 0.06 |
ENST00000358842.3 |
GK2 |
glycerol kinase 2 |
| chr2_+_1417228 | 0.06 |
ENST00000382269.3 ENST00000337415.3 ENST00000345913.4 ENST00000346956.3 ENST00000349624.3 ENST00000539820.1 ENST00000329066.4 ENST00000382201.3 |
TPO |
thyroid peroxidase |
| chr12_+_112204691 | 0.06 |
ENST00000416293.3 ENST00000261733.2 |
ALDH2 |
aldehyde dehydrogenase 2 family (mitochondrial) |
| chr15_+_81293254 | 0.06 |
ENST00000267984.2 |
MESDC1 |
mesoderm development candidate 1 |
| chr12_+_69979446 | 0.06 |
ENST00000543146.2 |
CCT2 |
chaperonin containing TCP1, subunit 2 (beta) |
| chr22_+_30821732 | 0.06 |
ENST00000355143.4 |
MTFP1 |
mitochondrial fission process 1 |
| chr10_+_70939983 | 0.06 |
ENST00000359655.4 ENST00000422378.1 |
SUPV3L1 |
suppressor of var1, 3-like 1 (S. cerevisiae) |
| chr15_+_62359175 | 0.05 |
ENST00000355522.5 |
C2CD4A |
C2 calcium-dependent domain containing 4A |
| chr2_-_99871570 | 0.05 |
ENST00000333017.2 ENST00000409679.1 ENST00000423306.1 |
LYG2 |
lysozyme G-like 2 |
| chr8_-_101962777 | 0.05 |
ENST00000395951.3 |
YWHAZ |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr19_-_55453077 | 0.05 |
ENST00000328092.5 ENST00000590030.1 |
NLRP7 |
NLR family, pyrin domain containing 7 |
| chr3_-_193272874 | 0.05 |
ENST00000342695.4 |
ATP13A4 |
ATPase type 13A4 |
| chr19_-_44008863 | 0.05 |
ENST00000601646.1 |
PHLDB3 |
pleckstrin homology-like domain, family B, member 3 |
| chr9_-_21368075 | 0.05 |
ENST00000449498.1 |
IFNA13 |
interferon, alpha 13 |
| chr1_+_110009215 | 0.05 |
ENST00000369872.3 |
SYPL2 |
synaptophysin-like 2 |
| chr22_+_39077947 | 0.05 |
ENST00000216034.4 |
TOMM22 |
translocase of outer mitochondrial membrane 22 homolog (yeast) |
| chr17_+_36584662 | 0.05 |
ENST00000431231.2 ENST00000437668.3 |
ARHGAP23 |
Rho GTPase activating protein 23 |
| chr8_-_8751068 | 0.05 |
ENST00000276282.6 |
MFHAS1 |
malignant fibrous histiocytoma amplified sequence 1 |
| chr12_-_75603482 | 0.05 |
ENST00000341669.3 ENST00000298972.1 ENST00000350228.2 |
KCNC2 |
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr1_-_53793725 | 0.05 |
ENST00000371454.2 |
LRP8 |
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr19_+_41256764 | 0.05 |
ENST00000243563.3 ENST00000601253.1 ENST00000597353.1 ENST00000599362.1 |
SNRPA |
small nuclear ribonucleoprotein polypeptide A |
| chr10_+_99332198 | 0.05 |
ENST00000307518.5 ENST00000298808.5 ENST00000370655.1 |
ANKRD2 |
ankyrin repeat domain 2 (stretch responsive muscle) |
| chr12_-_75603643 | 0.05 |
ENST00000549446.1 |
KCNC2 |
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr2_-_74781061 | 0.05 |
ENST00000264094.3 ENST00000393937.2 ENST00000409986.1 |
LOXL3 |
lysyl oxidase-like 3 |
| chr17_+_79679369 | 0.05 |
ENST00000350690.5 |
SLC25A10 |
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr2_+_177134201 | 0.05 |
ENST00000452865.1 |
MTX2 |
metaxin 2 |
| chr2_+_7017796 | 0.05 |
ENST00000382040.3 |
RSAD2 |
radical S-adenosyl methionine domain containing 2 |
| chr3_-_126327398 | 0.05 |
ENST00000383572.2 |
TXNRD3NB |
thioredoxin reductase 3 neighbor |
| chr9_+_113431059 | 0.05 |
ENST00000416899.2 |
MUSK |
muscle, skeletal, receptor tyrosine kinase |
| chr8_-_145047688 | 0.05 |
ENST00000356346.3 |
PLEC |
plectin |
| chr19_-_56826157 | 0.05 |
ENST00000592509.1 ENST00000592679.1 ENST00000588442.1 ENST00000593106.1 ENST00000587492.1 ENST00000254165.3 |
ZSCAN5A |
zinc finger and SCAN domain containing 5A |
| chr1_+_76540386 | 0.05 |
ENST00000328299.3 |
ST6GALNAC3 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
| chr12_+_49717081 | 0.05 |
ENST00000547807.1 ENST00000551567.1 |
TROAP |
trophinin associated protein |
| chr10_-_62332357 | 0.04 |
ENST00000503366.1 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
| chr22_+_29168652 | 0.04 |
ENST00000249064.4 ENST00000444523.1 ENST00000448492.2 ENST00000421503.2 |
CCDC117 |
coiled-coil domain containing 117 |
| chr1_-_6546001 | 0.04 |
ENST00000400913.1 |
PLEKHG5 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 5 |
| chr4_+_87515454 | 0.04 |
ENST00000427191.2 ENST00000436978.1 ENST00000502971.1 |
PTPN13 |
protein tyrosine phosphatase, non-receptor type 13 (APO-1/CD95 (Fas)-associated phosphatase) |
| chr15_-_31453157 | 0.04 |
ENST00000559177.1 ENST00000558445.1 |
TRPM1 |
transient receptor potential cation channel, subfamily M, member 1 |
| chr10_+_7745303 | 0.04 |
ENST00000429820.1 ENST00000379587.4 |
ITIH2 |
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr10_+_19777984 | 0.04 |
ENST00000377265.3 ENST00000455457.2 |
C10orf112 |
MAM and LDL receptor class A domain containing 1 |
| chr8_+_73921085 | 0.04 |
ENST00000276603.5 ENST00000276602.6 ENST00000518874.1 |
TERF1 |
telomeric repeat binding factor (NIMA-interacting) 1 |
| chr10_+_7745232 | 0.04 |
ENST00000358415.4 |
ITIH2 |
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr7_+_150076406 | 0.04 |
ENST00000329630.5 |
ZNF775 |
zinc finger protein 775 |
| chr7_+_95115210 | 0.04 |
ENST00000428113.1 ENST00000325885.5 |
ASB4 |
ankyrin repeat and SOCS box containing 4 |
| chr14_+_32798462 | 0.04 |
ENST00000280979.4 |
AKAP6 |
A kinase (PRKA) anchor protein 6 |
| chr2_+_11679963 | 0.04 |
ENST00000263834.5 |
GREB1 |
growth regulation by estrogen in breast cancer 1 |
| chr15_-_35280426 | 0.04 |
ENST00000559564.1 ENST00000356321.4 |
ZNF770 |
zinc finger protein 770 |
| chr7_-_19748640 | 0.04 |
ENST00000222567.5 |
TWISTNB |
TWIST neighbor |
| chr9_+_113431029 | 0.04 |
ENST00000189978.5 ENST00000374448.4 ENST00000374440.3 |
MUSK |
muscle, skeletal, receptor tyrosine kinase |
| chr8_-_124279627 | 0.04 |
ENST00000357082.4 |
ZHX1-C8ORF76 |
ZHX1-C8ORF76 readthrough |
| chr1_-_33502441 | 0.04 |
ENST00000548033.1 ENST00000487289.1 ENST00000373449.2 ENST00000480134.1 ENST00000467905.1 |
AK2 |
adenylate kinase 2 |
| chr11_-_8615507 | 0.04 |
ENST00000431279.2 ENST00000418597.1 |
STK33 |
serine/threonine kinase 33 |
| chr2_+_204801471 | 0.04 |
ENST00000316386.6 ENST00000435193.1 |
ICOS |
inducible T-cell co-stimulator |
| chr21_+_39628655 | 0.04 |
ENST00000398925.1 ENST00000398928.1 ENST00000328656.4 ENST00000443341.1 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr2_+_108994633 | 0.04 |
ENST00000409309.3 |
SULT1C4 |
sulfotransferase family, cytosolic, 1C, member 4 |
| chr13_+_30002741 | 0.04 |
ENST00000380808.2 |
MTUS2 |
microtubule associated tumor suppressor candidate 2 |
| chr11_-_64013288 | 0.04 |
ENST00000542235.1 |
PPP1R14B |
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr3_+_45984947 | 0.04 |
ENST00000304552.4 |
CXCR6 |
chemokine (C-X-C motif) receptor 6 |
| chr12_+_49717019 | 0.04 |
ENST00000549275.1 ENST00000551245.1 ENST00000380327.5 ENST00000548311.1 ENST00000550346.1 ENST00000550709.1 ENST00000549534.1 ENST00000257909.3 |
TROAP |
trophinin associated protein |
| chr2_-_208031542 | 0.04 |
ENST00000423015.1 |
KLF7 |
Kruppel-like factor 7 (ubiquitous) |
| chrX_+_100663243 | 0.04 |
ENST00000316594.5 |
HNRNPH2 |
heterogeneous nuclear ribonucleoprotein H2 (H') |
| chrX_+_148855726 | 0.04 |
ENST00000370416.4 |
HSFX1 |
heat shock transcription factor family, X linked 1 |
| chr4_+_71859156 | 0.04 |
ENST00000286648.5 ENST00000504730.1 ENST00000504952.1 |
DCK |
deoxycytidine kinase |
| chr17_-_40337470 | 0.04 |
ENST00000293330.1 |
HCRT |
hypocretin (orexin) neuropeptide precursor |
| chr1_+_180199393 | 0.04 |
ENST00000263726.2 |
LHX4 |
LIM homeobox 4 |
| chrX_-_148676974 | 0.03 |
ENST00000524178.1 |
HSFX2 |
heat shock transcription factor family, X linked 2 |
| chr12_-_67197760 | 0.03 |
ENST00000539540.1 ENST00000540433.1 ENST00000541947.1 ENST00000538373.1 |
GRIP1 |
glutamate receptor interacting protein 1 |
| chr2_+_234621551 | 0.03 |
ENST00000608381.1 ENST00000373414.3 |
UGT1A1 UGT1A5 |
UDP glucuronosyltransferase 1 family, polypeptide A8 UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr19_-_55690758 | 0.03 |
ENST00000590851.1 |
SYT5 |
synaptotagmin V |
| chr6_-_10435032 | 0.03 |
ENST00000491317.1 ENST00000496285.1 ENST00000479822.1 ENST00000487130.1 |
LINC00518 |
long intergenic non-protein coding RNA 518 |
| chr11_+_7506713 | 0.03 |
ENST00000329293.3 ENST00000534244.1 |
OLFML1 |
olfactomedin-like 1 |
| chr3_-_120068143 | 0.03 |
ENST00000295628.3 |
LRRC58 |
leucine rich repeat containing 58 |
| chr5_-_79287060 | 0.03 |
ENST00000512560.1 ENST00000509852.1 ENST00000512528.1 |
MTX3 |
metaxin 3 |
| chr17_+_79679299 | 0.03 |
ENST00000331531.5 |
SLC25A10 |
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr18_+_12407895 | 0.03 |
ENST00000590956.1 ENST00000336990.4 ENST00000440960.1 ENST00000588729.1 |
SLMO1 |
slowmo homolog 1 (Drosophila) |
| chrX_+_146993449 | 0.03 |
ENST00000218200.8 ENST00000370471.3 ENST00000370477.1 |
FMR1 |
fragile X mental retardation 1 |
| chr10_+_96522361 | 0.03 |
ENST00000371321.3 |
CYP2C19 |
cytochrome P450, family 2, subfamily C, polypeptide 19 |
| chr2_-_231989808 | 0.03 |
ENST00000258400.3 |
HTR2B |
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr2_-_29093132 | 0.03 |
ENST00000306108.5 |
TRMT61B |
tRNA methyltransferase 61 homolog B (S. cerevisiae) |
| chr20_-_656437 | 0.03 |
ENST00000488788.2 |
RP5-850E9.3 |
Uncharacterized protein |
| chr19_+_41257084 | 0.03 |
ENST00000601393.1 |
SNRPA |
small nuclear ribonucleoprotein polypeptide A |
| chr12_+_101988774 | 0.03 |
ENST00000545503.2 ENST00000536007.1 ENST00000541119.1 ENST00000361466.2 ENST00000551300.1 ENST00000550270.1 |
MYBPC1 |
myosin binding protein C, slow type |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0034136 | negative regulation of toll-like receptor 2 signaling pathway(GO:0034136) |
| 0.1 | 0.3 | GO:0032771 | regulation of monophenol monooxygenase activity(GO:0032771) positive regulation of monophenol monooxygenase activity(GO:0032773) negative regulation of catagen(GO:0051796) regulation of hair cycle by canonical Wnt signaling pathway(GO:0060901) positive regulation of melanosome transport(GO:1902910) |
| 0.1 | 0.2 | GO:0015766 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.1 | 0.2 | GO:0071810 | circadian temperature homeostasis(GO:0060086) regulation of fever generation by regulation of prostaglandin secretion(GO:0071810) positive regulation of fever generation by positive regulation of prostaglandin secretion(GO:0071812) positive regulation of ERK1 and ERK2 cascade via TNFSF11-mediated signaling(GO:0071848) regulation of fever generation by prostaglandin secretion(GO:0100009) |
| 0.1 | 0.2 | GO:0021718 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.1 | 0.2 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.1 | 0.3 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.0 | 0.2 | GO:0042335 | cuticle development(GO:0042335) |
| 0.0 | 0.1 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.0 | 0.1 | GO:0072560 | glandular epithelial cell maturation(GO:0002071) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) type B pancreatic cell maturation(GO:0072560) |
| 0.0 | 0.1 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.1 | GO:1904398 | positive regulation of neuromuscular junction development(GO:1904398) |
| 0.0 | 0.1 | GO:0015743 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.0 | 0.2 | GO:1903899 | lung goblet cell differentiation(GO:0060480) positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.0 | 0.2 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.1 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.0 | 0.1 | GO:0005989 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.0 | 0.1 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.0 | 0.1 | GO:0051086 | chaperone mediated protein folding independent of cofactor(GO:0051086) |
| 0.0 | 0.1 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.1 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) regulation of determination of dorsal identity(GO:2000015) |
| 0.0 | 0.1 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.1 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.0 | 0.0 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.1 | GO:0033504 | floor plate development(GO:0033504) |
| 0.0 | 0.0 | GO:1904815 | negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 0.0 | 0.1 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.1 | GO:2000051 | Wnt receptor catabolic process(GO:0038018) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.0 | 0.1 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.0 | 0.1 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.0 | 0.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.0 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 0.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.0 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 0.0 | 0.0 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.1 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.2 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.2 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 0.3 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.1 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.0 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.0 | 0.3 | GO:0032433 | filopodium tip(GO:0032433) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0008506 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 0.2 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.0 | 0.2 | GO:0035651 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.1 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.0 | 0.2 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.2 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.1 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) |
| 0.0 | 0.2 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.1 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.1 | GO:0000773 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.0 | 0.1 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.0 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.2 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.0 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) |
| 0.0 | 0.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.1 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.1 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0070404 | NADH binding(GO:0070404) |
| 0.0 | 0.0 | GO:0016429 | tRNA (adenine) methyltransferase activity(GO:0016426) tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |