Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
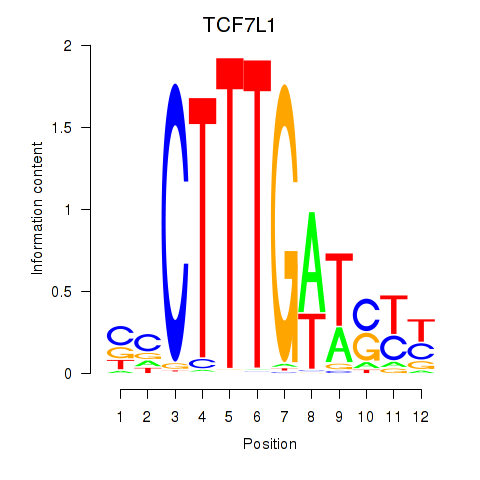
Results for TCF7L1
Z-value: 0.46
Transcription factors associated with TCF7L1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TCF7L1
|
ENSG00000152284.4 | TCF7L1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TCF7L1 | hg19_v2_chr2_+_85360499_85360598 | 0.83 | 1.1e-02 | Click! |
Activity profile of TCF7L1 motif
Sorted Z-values of TCF7L1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TCF7L1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_43885252 | 0.87 |
ENST00000453782.1 ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B |
creatine kinase, mitochondrial 1B |
| chr15_+_43985725 | 0.85 |
ENST00000413453.2 |
CKMT1A |
creatine kinase, mitochondrial 1A |
| chr15_+_43985084 | 0.84 |
ENST00000434505.1 ENST00000411750.1 |
CKMT1A |
creatine kinase, mitochondrial 1A |
| chr6_-_11779403 | 0.56 |
ENST00000414691.3 |
ADTRP |
androgen-dependent TFPI-regulating protein |
| chr5_+_66124590 | 0.54 |
ENST00000490016.2 ENST00000403666.1 ENST00000450827.1 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
| chr17_-_39507064 | 0.53 |
ENST00000007735.3 |
KRT33A |
keratin 33A |
| chr3_+_189349162 | 0.50 |
ENST00000264731.3 ENST00000382063.4 ENST00000418709.2 ENST00000320472.5 ENST00000392460.3 ENST00000440651.2 |
TP63 |
tumor protein p63 |
| chr6_-_11779174 | 0.48 |
ENST00000379413.2 |
ADTRP |
androgen-dependent TFPI-regulating protein |
| chr6_-_11779840 | 0.43 |
ENST00000506810.1 |
ADTRP |
androgen-dependent TFPI-regulating protein |
| chr11_-_87908600 | 0.40 |
ENST00000531138.1 ENST00000526372.1 ENST00000243662.6 |
RAB38 |
RAB38, member RAS oncogene family |
| chr5_-_175964366 | 0.39 |
ENST00000274811.4 |
RNF44 |
ring finger protein 44 |
| chr5_+_66300446 | 0.39 |
ENST00000261569.7 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
| chr15_+_41136216 | 0.33 |
ENST00000562057.1 ENST00000344051.4 |
SPINT1 |
serine peptidase inhibitor, Kunitz type 1 |
| chr9_-_23826298 | 0.32 |
ENST00000380117.1 |
ELAVL2 |
ELAV like neuron-specific RNA binding protein 2 |
| chrX_-_117119243 | 0.31 |
ENST00000539496.1 ENST00000469946.1 |
KLHL13 |
kelch-like family member 13 |
| chr17_+_30813576 | 0.30 |
ENST00000313401.3 |
CDK5R1 |
cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
| chrX_-_32173579 | 0.30 |
ENST00000359836.1 ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD |
dystrophin |
| chr14_-_23624511 | 0.29 |
ENST00000529705.2 |
SLC7A8 |
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chr7_-_25702669 | 0.28 |
ENST00000446840.1 |
AC003090.1 |
AC003090.1 |
| chr1_+_82266053 | 0.27 |
ENST00000370715.1 ENST00000370713.1 ENST00000319517.6 ENST00000370717.2 ENST00000394879.1 ENST00000271029.4 ENST00000335786.5 |
LPHN2 |
latrophilin 2 |
| chr14_+_75746781 | 0.26 |
ENST00000555347.1 |
FOS |
FBJ murine osteosarcoma viral oncogene homolog |
| chr1_-_109935819 | 0.26 |
ENST00000538502.1 |
SORT1 |
sortilin 1 |
| chr1_-_32801825 | 0.25 |
ENST00000329421.7 |
MARCKSL1 |
MARCKS-like 1 |
| chr5_-_16936340 | 0.24 |
ENST00000507288.1 ENST00000513610.1 |
MYO10 |
myosin X |
| chr10_+_71561649 | 0.24 |
ENST00000398978.3 ENST00000354547.3 ENST00000357811.3 |
COL13A1 |
collagen, type XIII, alpha 1 |
| chr8_-_42234745 | 0.24 |
ENST00000220812.2 |
DKK4 |
dickkopf WNT signaling pathway inhibitor 4 |
| chr1_-_6479963 | 0.24 |
ENST00000377836.4 ENST00000487437.1 ENST00000489730.1 ENST00000377834.4 |
HES2 |
hes family bHLH transcription factor 2 |
| chr10_+_71561630 | 0.23 |
ENST00000398974.3 ENST00000398971.3 ENST00000398968.3 ENST00000398966.3 ENST00000398964.3 ENST00000398969.3 ENST00000356340.3 ENST00000398972.3 ENST00000398973.3 |
COL13A1 |
collagen, type XIII, alpha 1 |
| chr3_+_132757215 | 0.21 |
ENST00000321871.6 ENST00000393130.3 ENST00000514894.1 ENST00000512662.1 |
TMEM108 |
transmembrane protein 108 |
| chr1_-_54303949 | 0.21 |
ENST00000234725.8 |
NDC1 |
NDC1 transmembrane nucleoporin |
| chr7_+_73245193 | 0.19 |
ENST00000340958.2 |
CLDN4 |
claudin 4 |
| chr8_+_32579341 | 0.19 |
ENST00000519240.1 ENST00000539990.1 |
NRG1 |
neuregulin 1 |
| chr1_-_54303934 | 0.19 |
ENST00000537333.1 |
NDC1 |
NDC1 transmembrane nucleoporin |
| chr12_-_48963829 | 0.19 |
ENST00000301046.2 ENST00000549817.1 |
LALBA |
lactalbumin, alpha- |
| chr2_-_9143786 | 0.18 |
ENST00000462696.1 ENST00000305997.3 |
MBOAT2 |
membrane bound O-acyltransferase domain containing 2 |
| chr18_+_11751493 | 0.18 |
ENST00000269162.5 |
GNAL |
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chrX_-_128788914 | 0.18 |
ENST00000429967.1 ENST00000307484.6 |
APLN |
apelin |
| chr2_-_208031542 | 0.18 |
ENST00000423015.1 |
KLF7 |
Kruppel-like factor 7 (ubiquitous) |
| chr4_-_103266355 | 0.18 |
ENST00000424970.2 |
SLC39A8 |
solute carrier family 39 (zinc transporter), member 8 |
| chr12_-_31477072 | 0.17 |
ENST00000454658.2 |
FAM60A |
family with sequence similarity 60, member A |
| chr12_+_70760056 | 0.17 |
ENST00000258111.4 |
KCNMB4 |
potassium large conductance calcium-activated channel, subfamily M, beta member 4 |
| chr6_-_107436473 | 0.16 |
ENST00000369042.1 |
BEND3 |
BEN domain containing 3 |
| chr11_-_117747434 | 0.16 |
ENST00000529335.2 ENST00000530956.1 ENST00000260282.4 |
FXYD6 |
FXYD domain containing ion transport regulator 6 |
| chr1_-_54304212 | 0.16 |
ENST00000540001.1 |
NDC1 |
NDC1 transmembrane nucleoporin |
| chr4_-_123542224 | 0.15 |
ENST00000264497.3 |
IL21 |
interleukin 21 |
| chr2_+_233734994 | 0.15 |
ENST00000331342.2 |
C2orf82 |
chromosome 2 open reading frame 82 |
| chr17_+_57297807 | 0.15 |
ENST00000284116.4 ENST00000581140.1 ENST00000581276.1 |
GDPD1 |
glycerophosphodiester phosphodiesterase domain containing 1 |
| chr15_-_52043722 | 0.14 |
ENST00000454181.2 |
LYSMD2 |
LysM, putative peptidoglycan-binding, domain containing 2 |
| chr22_-_32860427 | 0.14 |
ENST00000534972.1 ENST00000397450.1 ENST00000397452.1 |
BPIFC |
BPI fold containing family C |
| chr14_-_35344093 | 0.14 |
ENST00000382422.2 |
BAZ1A |
bromodomain adjacent to zinc finger domain, 1A |
| chr1_+_16085244 | 0.13 |
ENST00000400773.1 |
FBLIM1 |
filamin binding LIM protein 1 |
| chr10_+_71561704 | 0.13 |
ENST00000520267.1 |
COL13A1 |
collagen, type XIII, alpha 1 |
| chr4_+_95972822 | 0.13 |
ENST00000509540.1 ENST00000440890.2 |
BMPR1B |
bone morphogenetic protein receptor, type IB |
| chr1_-_53793584 | 0.12 |
ENST00000354412.3 ENST00000347547.2 ENST00000306052.6 |
LRP8 |
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr1_-_94079648 | 0.12 |
ENST00000370247.3 |
BCAR3 |
breast cancer anti-estrogen resistance 3 |
| chr1_-_116383322 | 0.12 |
ENST00000429731.1 |
NHLH2 |
nescient helix loop helix 2 |
| chr19_+_8274185 | 0.11 |
ENST00000558268.1 ENST00000558331.1 |
CERS4 |
ceramide synthase 4 |
| chr19_+_8274204 | 0.11 |
ENST00000561053.1 ENST00000251363.5 ENST00000559450.1 ENST00000559336.1 |
CERS4 |
ceramide synthase 4 |
| chr19_-_49864746 | 0.11 |
ENST00000598810.1 |
TEAD2 |
TEA domain family member 2 |
| chr14_-_23451845 | 0.11 |
ENST00000262713.2 |
AJUBA |
ajuba LIM protein |
| chr7_-_148725733 | 0.11 |
ENST00000286091.4 |
PDIA4 |
protein disulfide isomerase family A, member 4 |
| chr4_-_85419603 | 0.11 |
ENST00000295886.4 |
NKX6-1 |
NK6 homeobox 1 |
| chr17_-_37764128 | 0.11 |
ENST00000302584.4 |
NEUROD2 |
neuronal differentiation 2 |
| chr4_-_103266219 | 0.11 |
ENST00000394833.2 |
SLC39A8 |
solute carrier family 39 (zinc transporter), member 8 |
| chr1_-_177134024 | 0.11 |
ENST00000367654.3 |
ASTN1 |
astrotactin 1 |
| chr14_-_65409502 | 0.10 |
ENST00000389614.5 |
GPX2 |
glutathione peroxidase 2 (gastrointestinal) |
| chr6_+_96025341 | 0.10 |
ENST00000369293.1 ENST00000358812.4 |
MANEA |
mannosidase, endo-alpha |
| chr21_-_32185570 | 0.10 |
ENST00000329621.4 |
KRTAP8-1 |
keratin associated protein 8-1 |
| chr15_+_50474385 | 0.10 |
ENST00000267842.5 |
SLC27A2 |
solute carrier family 27 (fatty acid transporter), member 2 |
| chr1_-_43424500 | 0.10 |
ENST00000415851.2 ENST00000426263.3 ENST00000372500.3 |
SLC2A1 |
solute carrier family 2 (facilitated glucose transporter), member 1 |
| chr1_+_144339738 | 0.10 |
ENST00000538264.1 |
AL592284.1 |
Protein LOC642441 |
| chr3_+_123813543 | 0.10 |
ENST00000360013.3 |
KALRN |
kalirin, RhoGEF kinase |
| chr4_+_78078304 | 0.10 |
ENST00000316355.5 ENST00000354403.5 ENST00000502280.1 |
CCNG2 |
cyclin G2 |
| chr8_-_33424636 | 0.09 |
ENST00000256257.1 |
RNF122 |
ring finger protein 122 |
| chr1_-_53793725 | 0.09 |
ENST00000371454.2 |
LRP8 |
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr19_-_11450249 | 0.09 |
ENST00000222120.3 |
RAB3D |
RAB3D, member RAS oncogene family |
| chr4_-_120243545 | 0.09 |
ENST00000274024.3 |
FABP2 |
fatty acid binding protein 2, intestinal |
| chr2_-_208031943 | 0.09 |
ENST00000421199.1 ENST00000457962.1 |
KLF7 |
Kruppel-like factor 7 (ubiquitous) |
| chr10_+_71562180 | 0.09 |
ENST00000517713.1 ENST00000522165.1 ENST00000520133.1 |
COL13A1 |
collagen, type XIII, alpha 1 |
| chr19_+_11071652 | 0.09 |
ENST00000344626.4 ENST00000429416.3 |
SMARCA4 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr7_+_139528952 | 0.08 |
ENST00000416849.2 ENST00000436047.2 ENST00000414508.2 ENST00000448866.1 |
TBXAS1 |
thromboxane A synthase 1 (platelet) |
| chr7_+_115850547 | 0.08 |
ENST00000358204.4 ENST00000455989.1 ENST00000537767.1 |
TES |
testis derived transcript (3 LIM domains) |
| chr8_+_29953163 | 0.08 |
ENST00000518192.1 |
LEPROTL1 |
leptin receptor overlapping transcript-like 1 |
| chr15_+_81293254 | 0.08 |
ENST00000267984.2 |
MESDC1 |
mesoderm development candidate 1 |
| chr2_+_16080659 | 0.08 |
ENST00000281043.3 |
MYCN |
v-myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog |
| chr13_-_72440901 | 0.08 |
ENST00000359684.2 |
DACH1 |
dachshund homolog 1 (Drosophila) |
| chr17_-_7297833 | 0.08 |
ENST00000571802.1 ENST00000576201.1 ENST00000573213.1 ENST00000324822.11 |
TMEM256-PLSCR3 |
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr17_-_46035187 | 0.07 |
ENST00000300557.2 |
PRR15L |
proline rich 15-like |
| chr17_+_56315787 | 0.07 |
ENST00000262290.4 ENST00000421678.2 |
LPO |
lactoperoxidase |
| chr19_+_11071546 | 0.07 |
ENST00000358026.2 |
SMARCA4 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr6_+_143929307 | 0.07 |
ENST00000427704.2 ENST00000305766.6 |
PHACTR2 |
phosphatase and actin regulator 2 |
| chr12_+_56324933 | 0.07 |
ENST00000549629.1 ENST00000555218.1 |
DGKA |
diacylglycerol kinase, alpha 80kDa |
| chr10_-_101380121 | 0.07 |
ENST00000370495.4 |
SLC25A28 |
solute carrier family 25 (mitochondrial iron transporter), member 28 |
| chr6_-_29527702 | 0.07 |
ENST00000377050.4 |
UBD |
ubiquitin D |
| chr4_+_113152881 | 0.07 |
ENST00000274000.5 |
AP1AR |
adaptor-related protein complex 1 associated regulatory protein |
| chr1_-_177133818 | 0.07 |
ENST00000424564.2 ENST00000361833.2 |
ASTN1 |
astrotactin 1 |
| chr3_+_52813932 | 0.07 |
ENST00000537050.1 |
ITIH1 |
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr12_+_56324756 | 0.07 |
ENST00000331886.5 ENST00000555090.1 |
DGKA |
diacylglycerol kinase, alpha 80kDa |
| chr21_-_34143971 | 0.07 |
ENST00000290178.4 |
PAXBP1 |
PAX3 and PAX7 binding protein 1 |
| chr7_+_128828713 | 0.07 |
ENST00000249373.3 |
SMO |
smoothened, frizzled family receptor |
| chr8_-_16859690 | 0.07 |
ENST00000180166.5 |
FGF20 |
fibroblast growth factor 20 |
| chr13_+_30002741 | 0.07 |
ENST00000380808.2 |
MTUS2 |
microtubule associated tumor suppressor candidate 2 |
| chr17_-_48072574 | 0.07 |
ENST00000434704.2 |
DLX3 |
distal-less homeobox 3 |
| chr17_-_39093672 | 0.07 |
ENST00000209718.3 ENST00000436344.3 ENST00000485751.1 |
KRT23 |
keratin 23 (histone deacetylase inducible) |
| chr8_+_29952914 | 0.06 |
ENST00000321250.8 ENST00000518001.1 ENST00000520682.1 ENST00000442880.2 ENST00000523116.1 |
LEPROTL1 |
leptin receptor overlapping transcript-like 1 |
| chr17_-_10421853 | 0.06 |
ENST00000226207.5 |
MYH1 |
myosin, heavy chain 1, skeletal muscle, adult |
| chr6_-_33548006 | 0.06 |
ENST00000374467.3 |
BAK1 |
BCL2-antagonist/killer 1 |
| chr3_+_179280668 | 0.06 |
ENST00000429709.2 ENST00000450518.2 ENST00000392662.1 ENST00000490364.1 |
ACTL6A |
actin-like 6A |
| chr2_-_241725781 | 0.06 |
ENST00000428768.1 |
KIF1A |
kinesin family member 1A |
| chr11_-_94965667 | 0.06 |
ENST00000542176.1 ENST00000278499.2 |
SESN3 |
sestrin 3 |
| chr15_-_65670360 | 0.06 |
ENST00000327987.4 |
IGDCC3 |
immunoglobulin superfamily, DCC subclass, member 3 |
| chr19_+_41256764 | 0.06 |
ENST00000243563.3 ENST00000601253.1 ENST00000597353.1 ENST00000599362.1 |
SNRPA |
small nuclear ribonucleoprotein polypeptide A |
| chr2_-_80531399 | 0.06 |
ENST00000409148.1 ENST00000415098.1 ENST00000452811.1 |
LRRTM1 |
leucine rich repeat transmembrane neuronal 1 |
| chr6_-_33547975 | 0.06 |
ENST00000442998.2 ENST00000360661.5 |
BAK1 |
BCL2-antagonist/killer 1 |
| chr3_-_57199397 | 0.06 |
ENST00000296318.7 |
IL17RD |
interleukin 17 receptor D |
| chr20_+_18794370 | 0.06 |
ENST00000377428.2 |
SCP2D1 |
SCP2 sterol-binding domain containing 1 |
| chr17_-_7297519 | 0.06 |
ENST00000576362.1 ENST00000571078.1 |
TMEM256-PLSCR3 |
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr2_+_171571827 | 0.06 |
ENST00000375281.3 |
SP5 |
Sp5 transcription factor |
| chr11_+_36317830 | 0.05 |
ENST00000530639.1 |
PRR5L |
proline rich 5 like |
| chr17_+_80416050 | 0.05 |
ENST00000579198.1 ENST00000390006.4 ENST00000580296.1 |
NARF |
nuclear prelamin A recognition factor |
| chr14_-_21516590 | 0.05 |
ENST00000555026.1 |
NDRG2 |
NDRG family member 2 |
| chr9_+_116225999 | 0.05 |
ENST00000317613.6 |
RGS3 |
regulator of G-protein signaling 3 |
| chr14_-_23451467 | 0.05 |
ENST00000555074.1 ENST00000361265.4 |
RP11-298I3.5 AJUBA |
RP11-298I3.5 ajuba LIM protein |
| chr6_-_32083106 | 0.05 |
ENST00000442721.1 |
TNXB |
tenascin XB |
| chr2_+_9983483 | 0.05 |
ENST00000263663.5 |
TAF1B |
TATA box binding protein (TBP)-associated factor, RNA polymerase I, B, 63kDa |
| chr17_+_56315936 | 0.05 |
ENST00000543544.1 |
LPO |
lactoperoxidase |
| chr2_+_234686976 | 0.05 |
ENST00000389758.3 |
MROH2A |
maestro heat-like repeat family member 2A |
| chr21_-_46330545 | 0.05 |
ENST00000320216.6 ENST00000397852.1 |
ITGB2 |
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr1_+_180199393 | 0.05 |
ENST00000263726.2 |
LHX4 |
LIM homeobox 4 |
| chr19_+_51152702 | 0.05 |
ENST00000425202.1 |
C19orf81 |
chromosome 19 open reading frame 81 |
| chr3_-_148939835 | 0.05 |
ENST00000264613.6 |
CP |
ceruloplasmin (ferroxidase) |
| chr10_-_126107482 | 0.05 |
ENST00000368845.5 ENST00000539214.1 |
OAT |
ornithine aminotransferase |
| chr20_+_55966444 | 0.05 |
ENST00000356208.5 ENST00000440234.2 |
RBM38 |
RNA binding motif protein 38 |
| chr16_+_19222479 | 0.05 |
ENST00000568433.1 |
SYT17 |
synaptotagmin XVII |
| chr5_+_149546334 | 0.05 |
ENST00000231656.8 |
CDX1 |
caudal type homeobox 1 |
| chr15_+_75074410 | 0.05 |
ENST00000439220.2 |
CSK |
c-src tyrosine kinase |
| chrX_-_110655391 | 0.05 |
ENST00000356915.2 ENST00000356220.3 |
DCX |
doublecortin |
| chr4_+_113152978 | 0.05 |
ENST00000309703.6 |
AP1AR |
adaptor-related protein complex 1 associated regulatory protein |
| chr13_-_39612176 | 0.04 |
ENST00000352251.3 ENST00000350125.3 |
PROSER1 |
proline and serine rich 1 |
| chr2_+_48796120 | 0.04 |
ENST00000394754.1 |
STON1-GTF2A1L |
STON1-GTF2A1L readthrough |
| chr11_-_117747327 | 0.04 |
ENST00000584230.1 ENST00000527429.1 ENST00000584394.1 ENST00000532984.1 |
FXYD6 FXYD6-FXYD2 |
FXYD domain containing ion transport regulator 6 FXYD6-FXYD2 readthrough |
| chr2_-_39348137 | 0.04 |
ENST00000426016.1 |
SOS1 |
son of sevenless homolog 1 (Drosophila) |
| chr19_+_41257084 | 0.04 |
ENST00000601393.1 |
SNRPA |
small nuclear ribonucleoprotein polypeptide A |
| chr1_-_115053781 | 0.04 |
ENST00000358465.2 ENST00000369543.2 |
TRIM33 |
tripartite motif containing 33 |
| chr14_+_39644387 | 0.04 |
ENST00000553331.1 ENST00000216832.4 |
PNN |
pinin, desmosome associated protein |
| chrX_-_110655306 | 0.04 |
ENST00000371993.2 |
DCX |
doublecortin |
| chr1_+_212797789 | 0.04 |
ENST00000294829.3 |
FAM71A |
family with sequence similarity 71, member A |
| chr6_-_32145861 | 0.04 |
ENST00000336984.6 |
AGPAT1 |
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr2_+_9983642 | 0.04 |
ENST00000396242.3 |
TAF1B |
TATA box binding protein (TBP)-associated factor, RNA polymerase I, B, 63kDa |
| chr17_+_35851570 | 0.04 |
ENST00000394386.1 |
DUSP14 |
dual specificity phosphatase 14 |
| chr7_+_29234101 | 0.04 |
ENST00000435288.2 |
CHN2 |
chimerin 2 |
| chr2_-_100721923 | 0.04 |
ENST00000356421.2 |
AFF3 |
AF4/FMR2 family, member 3 |
| chr10_+_11206925 | 0.04 |
ENST00000354440.2 ENST00000315874.4 ENST00000427450.1 |
CELF2 |
CUGBP, Elav-like family member 2 |
| chr10_+_114710211 | 0.03 |
ENST00000349937.2 ENST00000369397.4 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr15_+_65134088 | 0.03 |
ENST00000323544.4 ENST00000437723.1 |
PLEKHO2 AC069368.3 |
pleckstrin homology domain containing, family O member 2 Uncharacterized protein |
| chr17_+_57232690 | 0.03 |
ENST00000262293.4 |
PRR11 |
proline rich 11 |
| chr17_-_10372875 | 0.03 |
ENST00000255381.2 |
MYH4 |
myosin, heavy chain 4, skeletal muscle |
| chr2_+_234621551 | 0.03 |
ENST00000608381.1 ENST00000373414.3 |
UGT1A1 UGT1A5 |
UDP glucuronosyltransferase 1 family, polypeptide A8 UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr11_-_65381643 | 0.03 |
ENST00000309100.3 ENST00000529839.1 ENST00000526293.1 |
MAP3K11 |
mitogen-activated protein kinase kinase kinase 11 |
| chr21_-_34144157 | 0.03 |
ENST00000331923.4 |
PAXBP1 |
PAX3 and PAX7 binding protein 1 |
| chr7_-_27219849 | 0.03 |
ENST00000396344.4 |
HOXA10 |
homeobox A10 |
| chr6_+_134274322 | 0.03 |
ENST00000367871.1 ENST00000237264.4 |
TBPL1 |
TBP-like 1 |
| chr1_+_160160283 | 0.03 |
ENST00000368079.3 |
CASQ1 |
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr17_+_59529743 | 0.03 |
ENST00000589003.1 ENST00000393853.4 |
TBX4 |
T-box 4 |
| chr1_+_241815577 | 0.03 |
ENST00000366552.2 ENST00000437684.2 |
WDR64 |
WD repeat domain 64 |
| chr15_+_80696666 | 0.03 |
ENST00000303329.4 |
ARNT2 |
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr10_-_99052382 | 0.03 |
ENST00000453547.2 ENST00000316676.8 ENST00000358308.3 ENST00000466484.1 ENST00000358531.4 |
ARHGAP19-SLIT1 ARHGAP19 |
ARHGAP19-SLIT1 readthrough (NMD candidate) Rho GTPase activating protein 19 |
| chr9_-_74979420 | 0.03 |
ENST00000343431.2 ENST00000376956.3 |
ZFAND5 |
zinc finger, AN1-type domain 5 |
| chr9_-_34381536 | 0.03 |
ENST00000379126.3 ENST00000379127.1 ENST00000379133.3 |
C9orf24 |
chromosome 9 open reading frame 24 |
| chr7_+_29234028 | 0.03 |
ENST00000222792.6 |
CHN2 |
chimerin 2 |
| chr20_-_656437 | 0.03 |
ENST00000488788.2 |
RP5-850E9.3 |
Uncharacterized protein |
| chr4_+_79567314 | 0.03 |
ENST00000503539.1 ENST00000504675.1 |
RP11-792D21.2 |
long intergenic non-protein coding RNA 1094 |
| chr4_-_140544386 | 0.03 |
ENST00000561977.1 |
RP11-308D13.3 |
RP11-308D13.3 |
| chr19_-_2328572 | 0.02 |
ENST00000252622.10 |
LSM7 |
LSM7 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr17_+_7533439 | 0.02 |
ENST00000441599.2 ENST00000380450.4 ENST00000416273.3 ENST00000575903.1 ENST00000576830.1 ENST00000571153.1 ENST00000575618.1 ENST00000576152.1 |
SHBG |
sex hormone-binding globulin |
| chr5_-_160973649 | 0.02 |
ENST00000393959.1 ENST00000517547.1 |
GABRB2 |
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr3_-_160283348 | 0.02 |
ENST00000334256.4 |
KPNA4 |
karyopherin alpha 4 (importin alpha 3) |
| chr21_+_18811205 | 0.02 |
ENST00000440664.1 |
C21orf37 |
chromosome 21 open reading frame 37 |
| chr18_+_3449821 | 0.02 |
ENST00000407501.2 ENST00000405385.3 ENST00000546979.1 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr14_+_57857262 | 0.02 |
ENST00000555166.1 ENST00000556492.1 ENST00000554703.1 |
NAA30 |
N(alpha)-acetyltransferase 30, NatC catalytic subunit |
| chr12_+_53491220 | 0.02 |
ENST00000548547.1 ENST00000301464.3 |
IGFBP6 |
insulin-like growth factor binding protein 6 |
| chr9_-_88896977 | 0.02 |
ENST00000311534.6 |
ISCA1 |
iron-sulfur cluster assembly 1 |
| chr11_+_77184416 | 0.02 |
ENST00000598970.1 |
DKFZP434E1119 |
DKFZP434E1119 |
| chr4_-_146859623 | 0.02 |
ENST00000379448.4 ENST00000513320.1 |
ZNF827 |
zinc finger protein 827 |
| chr16_-_18812746 | 0.02 |
ENST00000546206.2 ENST00000562819.1 ENST00000562234.2 ENST00000304414.7 ENST00000567078.2 |
ARL6IP1 RP11-1035H13.3 |
ADP-ribosylation factor-like 6 interacting protein 1 Uncharacterized protein |
| chr9_-_34381511 | 0.02 |
ENST00000379124.1 |
C9orf24 |
chromosome 9 open reading frame 24 |
| chr3_+_123813509 | 0.02 |
ENST00000460856.1 ENST00000240874.3 |
KALRN |
kalirin, RhoGEF kinase |
| chr6_+_35704804 | 0.02 |
ENST00000373869.3 |
ARMC12 |
armadillo repeat containing 12 |
| chr20_-_17511962 | 0.02 |
ENST00000377873.3 |
BFSP1 |
beaded filament structural protein 1, filensin |
| chr6_+_35704855 | 0.02 |
ENST00000288065.2 ENST00000373866.3 |
ARMC12 |
armadillo repeat containing 12 |
| chr8_-_119964434 | 0.02 |
ENST00000297350.4 |
TNFRSF11B |
tumor necrosis factor receptor superfamily, member 11b |
| chr7_+_73868439 | 0.02 |
ENST00000424337.2 |
GTF2IRD1 |
GTF2I repeat domain containing 1 |
| chr2_+_179184955 | 0.02 |
ENST00000315022.2 |
OSBPL6 |
oxysterol binding protein-like 6 |
| chr8_-_70745575 | 0.02 |
ENST00000524945.1 |
SLCO5A1 |
solute carrier organic anion transporter family, member 5A1 |
| chr10_+_7745303 | 0.02 |
ENST00000429820.1 ENST00000379587.4 |
ITIH2 |
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr4_-_139163491 | 0.02 |
ENST00000280612.5 |
SLC7A11 |
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
| chr5_-_148930960 | 0.02 |
ENST00000261798.5 ENST00000377843.2 |
CSNK1A1 |
casein kinase 1, alpha 1 |
| chrX_+_69488155 | 0.02 |
ENST00000374495.3 |
ARR3 |
arrestin 3, retinal (X-arrestin) |
| chr10_+_7745232 | 0.02 |
ENST00000358415.4 |
ITIH2 |
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr4_+_79567362 | 0.02 |
ENST00000512322.1 |
RP11-792D21.2 |
long intergenic non-protein coding RNA 1094 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0071789 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.2 | 1.7 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.1 | 0.5 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.1 | 0.3 | GO:0021722 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.1 | 0.3 | GO:0005989 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.1 | 0.2 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.1 | 0.4 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.3 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.0 | 0.1 | GO:0002352 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 | 0.1 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.0 | 0.1 | GO:0072560 | glandular epithelial cell maturation(GO:0002071) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) type B pancreatic cell maturation(GO:0072560) |
| 0.0 | 0.1 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.2 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.3 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.2 | GO:0007068 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 0.0 | 0.3 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.2 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.2 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.1 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.0 | 0.3 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.3 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.1 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.1 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.0 | 0.1 | GO:0001189 | RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.0 | 0.2 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.1 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.1 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 0.1 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 0.3 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.7 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.2 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.2 | 0.7 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.1 | 0.4 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.1 | 0.3 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.3 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.1 | GO:0005668 | RNA polymerase transcription factor SL1 complex(GO:0005668) |
| 0.0 | 0.3 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.1 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.1 | GO:0001939 | female pronucleus(GO:0001939) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.5 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.1 | 0.3 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.1 | 0.2 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.1 | 0.3 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.0 | 0.2 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.5 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.0 | 0.2 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.3 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.6 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.2 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 0.3 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.2 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.3 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.2 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.7 | NABA COLLAGENS | Genes encoding collagen proteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |