Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for TFAP4_MSC
Z-value: 0.85
Transcription factors associated with TFAP4_MSC
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
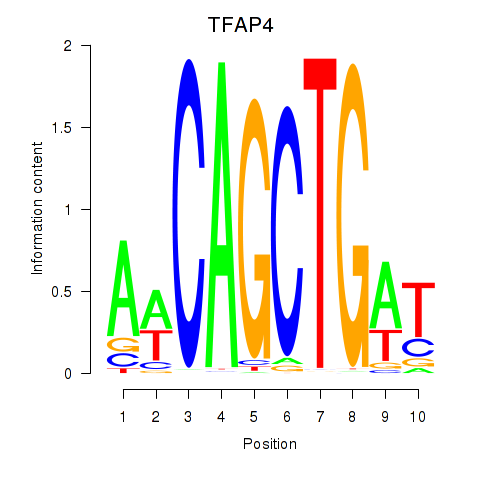
TFAP4
|
ENSG00000090447.7 | TFAP4 |
|
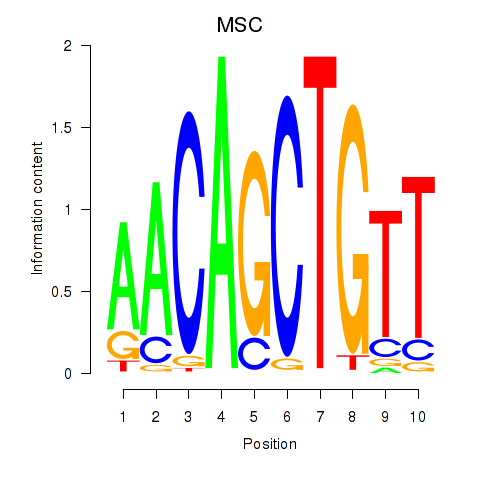
MSC
|
ENSG00000178860.8 | MSC |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MSC | hg19_v2_chr8_-_72756667_72756736 | 0.84 | 9.0e-03 | Click! |
| TFAP4 | hg19_v2_chr16_-_4323015_4323076 | -0.65 | 8.0e-02 | Click! |
Activity profile of TFAP4_MSC motif
Sorted Z-values of TFAP4_MSC motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TFAP4_MSC
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_225811747 | 1.53 |
ENST00000409592.3 |
DOCK10 |
dedicator of cytokinesis 10 |
| chr6_-_31697255 | 1.52 |
ENST00000436437.1 |
DDAH2 |
dimethylarginine dimethylaminohydrolase 2 |
| chr13_+_102104980 | 1.51 |
ENST00000545560.2 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr17_-_53800217 | 1.36 |
ENST00000424486.2 |
TMEM100 |
transmembrane protein 100 |
| chr13_+_102104952 | 1.33 |
ENST00000376180.3 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr20_+_33292068 | 1.23 |
ENST00000374810.3 ENST00000374809.2 ENST00000451665.1 |
TP53INP2 |
tumor protein p53 inducible nuclear protein 2 |
| chr3_-_178790057 | 1.12 |
ENST00000311417.2 |
ZMAT3 |
zinc finger, matrin-type 3 |
| chr5_+_95066823 | 1.10 |
ENST00000506817.1 ENST00000379982.3 |
RHOBTB3 |
Rho-related BTB domain containing 3 |
| chr3_-_178789220 | 1.07 |
ENST00000414084.1 |
ZMAT3 |
zinc finger, matrin-type 3 |
| chr6_-_31697563 | 1.03 |
ENST00000375789.2 ENST00000416410.1 |
DDAH2 |
dimethylarginine dimethylaminohydrolase 2 |
| chr8_-_95961578 | 0.85 |
ENST00000448464.2 ENST00000342697.4 |
TP53INP1 |
tumor protein p53 inducible nuclear protein 1 |
| chr19_+_18208603 | 0.79 |
ENST00000262811.6 |
MAST3 |
microtubule associated serine/threonine kinase 3 |
| chr11_-_111783595 | 0.71 |
ENST00000528628.1 |
CRYAB |
crystallin, alpha B |
| chr8_-_13134045 | 0.71 |
ENST00000512044.2 |
DLC1 |
deleted in liver cancer 1 |
| chr1_+_223101757 | 0.70 |
ENST00000284476.6 |
DISP1 |
dispatched homolog 1 (Drosophila) |
| chr1_+_78354297 | 0.64 |
ENST00000334785.7 |
NEXN |
nexilin (F actin binding protein) |
| chr6_-_24911195 | 0.64 |
ENST00000259698.4 |
FAM65B |
family with sequence similarity 65, member B |
| chr11_-_70507901 | 0.62 |
ENST00000449833.2 ENST00000357171.3 ENST00000449116.2 |
SHANK2 |
SH3 and multiple ankyrin repeat domains 2 |
| chr22_+_31003133 | 0.61 |
ENST00000405742.3 |
TCN2 |
transcobalamin II |
| chr17_-_7145106 | 0.60 |
ENST00000577035.1 |
GABARAP |
GABA(A) receptor-associated protein |
| chr2_-_190044480 | 0.60 |
ENST00000374866.3 |
COL5A2 |
collagen, type V, alpha 2 |
| chr13_-_33859819 | 0.59 |
ENST00000336934.5 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
| chr6_-_117747015 | 0.59 |
ENST00000368508.3 ENST00000368507.3 |
ROS1 |
c-ros oncogene 1 , receptor tyrosine kinase |
| chr15_-_52944231 | 0.59 |
ENST00000546305.2 |
FAM214A |
family with sequence similarity 214, member A |
| chr4_-_89619386 | 0.58 |
ENST00000323061.5 |
NAP1L5 |
nucleosome assembly protein 1-like 5 |
| chr22_+_35776828 | 0.58 |
ENST00000216117.8 |
HMOX1 |
heme oxygenase (decycling) 1 |
| chr12_-_54121212 | 0.58 |
ENST00000548263.1 ENST00000430117.2 ENST00000550804.1 ENST00000549173.1 ENST00000551900.1 ENST00000546619.1 ENST00000548177.1 ENST00000549349.1 |
CALCOCO1 |
calcium binding and coiled-coil domain 1 |
| chr1_+_78354175 | 0.57 |
ENST00000401035.3 ENST00000457030.1 ENST00000330010.8 |
NEXN |
nexilin (F actin binding protein) |
| chr22_+_31002779 | 0.57 |
ENST00000215838.3 |
TCN2 |
transcobalamin II |
| chr12_-_54121261 | 0.55 |
ENST00000549784.1 ENST00000262059.4 |
CALCOCO1 |
calcium binding and coiled-coil domain 1 |
| chr16_-_31076332 | 0.54 |
ENST00000539836.3 ENST00000535577.1 ENST00000442862.2 |
ZNF668 |
zinc finger protein 668 |
| chr13_-_43566301 | 0.54 |
ENST00000398762.3 ENST00000313640.7 ENST00000313624.7 |
EPSTI1 |
epithelial stromal interaction 1 (breast) |
| chr5_-_95158644 | 0.54 |
ENST00000237858.6 |
GLRX |
glutaredoxin (thioltransferase) |
| chr12_-_56236734 | 0.53 |
ENST00000548629.1 |
MMP19 |
matrix metallopeptidase 19 |
| chr6_+_155470243 | 0.53 |
ENST00000456877.2 ENST00000528391.2 |
TIAM2 |
T-cell lymphoma invasion and metastasis 2 |
| chr6_+_155537771 | 0.52 |
ENST00000275246.7 |
TIAM2 |
T-cell lymphoma invasion and metastasis 2 |
| chr1_-_42384343 | 0.52 |
ENST00000372584.1 |
HIVEP3 |
human immunodeficiency virus type I enhancer binding protein 3 |
| chr10_-_102089729 | 0.51 |
ENST00000465680.2 |
PKD2L1 |
polycystic kidney disease 2-like 1 |
| chr1_-_86043921 | 0.48 |
ENST00000535924.2 |
DDAH1 |
dimethylarginine dimethylaminohydrolase 1 |
| chr9_-_123239632 | 0.48 |
ENST00000416449.1 |
CDK5RAP2 |
CDK5 regulatory subunit associated protein 2 |
| chr20_+_62697564 | 0.47 |
ENST00000458442.1 |
TCEA2 |
transcription elongation factor A (SII), 2 |
| chrX_-_107019181 | 0.46 |
ENST00000315660.4 ENST00000372384.2 ENST00000502650.1 ENST00000506724.1 |
TSC22D3 |
TSC22 domain family, member 3 |
| chr2_-_175629135 | 0.45 |
ENST00000409542.1 ENST00000409219.1 |
CHRNA1 |
cholinergic receptor, nicotinic, alpha 1 (muscle) |
| chr9_+_17134980 | 0.44 |
ENST00000380647.3 |
CNTLN |
centlein, centrosomal protein |
| chr1_-_243326612 | 0.43 |
ENST00000492145.1 ENST00000490813.1 ENST00000464936.1 |
CEP170 |
centrosomal protein 170kDa |
| chr12_-_108733078 | 0.43 |
ENST00000552995.1 ENST00000312143.7 ENST00000397688.2 ENST00000550402.1 |
CMKLR1 |
chemokine-like receptor 1 |
| chr2_-_128400788 | 0.42 |
ENST00000409286.1 |
LIMS2 |
LIM and senescent cell antigen-like domains 2 |
| chr16_-_31105870 | 0.41 |
ENST00000394971.3 |
VKORC1 |
vitamin K epoxide reductase complex, subunit 1 |
| chr2_+_30370382 | 0.41 |
ENST00000402708.1 |
YPEL5 |
yippee-like 5 (Drosophila) |
| chr9_+_131174024 | 0.39 |
ENST00000420034.1 ENST00000372842.1 |
CERCAM |
cerebral endothelial cell adhesion molecule |
| chr16_-_31106048 | 0.39 |
ENST00000300851.6 |
VKORC1 |
vitamin K epoxide reductase complex, subunit 1 |
| chr2_+_54198210 | 0.39 |
ENST00000607452.1 ENST00000422521.2 |
ACYP2 |
acylphosphatase 2, muscle type |
| chr14_+_24584508 | 0.39 |
ENST00000559354.1 ENST00000560459.1 ENST00000559593.1 ENST00000396941.4 ENST00000396936.1 |
DCAF11 |
DDB1 and CUL4 associated factor 11 |
| chr1_+_159750776 | 0.39 |
ENST00000368107.1 |
DUSP23 |
dual specificity phosphatase 23 |
| chr10_+_1102721 | 0.38 |
ENST00000263150.4 |
WDR37 |
WD repeat domain 37 |
| chr16_-_31106211 | 0.38 |
ENST00000532364.1 ENST00000529564.1 ENST00000319788.7 ENST00000354895.4 ENST00000394975.2 |
RP11-196G11.1 VKORC1 |
Uncharacterized protein vitamin K epoxide reductase complex, subunit 1 |
| chr22_+_31003190 | 0.38 |
ENST00000407817.3 |
TCN2 |
transcobalamin II |
| chrX_-_150067173 | 0.38 |
ENST00000370377.3 ENST00000320893.6 |
CD99L2 |
CD99 molecule-like 2 |
| chr1_+_159750720 | 0.38 |
ENST00000368109.1 ENST00000368108.3 |
DUSP23 |
dual specificity phosphatase 23 |
| chr1_-_161993422 | 0.37 |
ENST00000367940.2 |
OLFML2B |
olfactomedin-like 2B |
| chr19_+_35521572 | 0.36 |
ENST00000262631.5 |
SCN1B |
sodium channel, voltage-gated, type I, beta subunit |
| chr8_+_77593448 | 0.36 |
ENST00000521891.2 |
ZFHX4 |
zinc finger homeobox 4 |
| chr8_-_72274095 | 0.36 |
ENST00000303824.7 |
EYA1 |
eyes absent homolog 1 (Drosophila) |
| chr2_+_56411131 | 0.36 |
ENST00000407595.2 |
CCDC85A |
coiled-coil domain containing 85A |
| chr11_-_2160180 | 0.36 |
ENST00000381406.4 |
IGF2 |
insulin-like growth factor 2 (somatomedin A) |
| chr9_-_104357277 | 0.35 |
ENST00000374806.1 |
PPP3R2 |
protein phosphatase 3, regulatory subunit B, beta |
| chr16_-_31076273 | 0.35 |
ENST00000426488.2 |
ZNF668 |
zinc finger protein 668 |
| chr11_+_33061543 | 0.34 |
ENST00000432887.1 ENST00000528898.1 ENST00000531632.2 |
TCP11L1 |
t-complex 11, testis-specific-like 1 |
| chr2_+_148778570 | 0.34 |
ENST00000407073.1 |
MBD5 |
methyl-CpG binding domain protein 5 |
| chr10_-_64576105 | 0.34 |
ENST00000242480.3 ENST00000411732.1 |
EGR2 |
early growth response 2 |
| chr2_-_158345462 | 0.34 |
ENST00000439355.1 ENST00000540637.1 |
CYTIP |
cytohesin 1 interacting protein |
| chr11_+_71938925 | 0.34 |
ENST00000538751.1 |
INPPL1 |
inositol polyphosphate phosphatase-like 1 |
| chr15_+_93447675 | 0.34 |
ENST00000536619.1 |
CHD2 |
chromodomain helicase DNA binding protein 2 |
| chr8_+_77593474 | 0.33 |
ENST00000455469.2 ENST00000050961.6 |
ZFHX4 |
zinc finger homeobox 4 |
| chr5_+_49962772 | 0.33 |
ENST00000281631.5 ENST00000513738.1 ENST00000503665.1 ENST00000514067.2 ENST00000503046.1 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
| chr19_-_46285646 | 0.32 |
ENST00000458663.2 |
DMPK |
dystrophia myotonica-protein kinase |
| chr11_+_45918092 | 0.32 |
ENST00000395629.2 |
MAPK8IP1 |
mitogen-activated protein kinase 8 interacting protein 1 |
| chr2_+_95537170 | 0.32 |
ENST00000295201.4 |
TEKT4 |
tektin 4 |
| chr13_+_76362974 | 0.32 |
ENST00000497947.2 |
LMO7 |
LIM domain 7 |
| chr14_-_73493784 | 0.31 |
ENST00000553891.1 |
ZFYVE1 |
zinc finger, FYVE domain containing 1 |
| chr3_-_81792780 | 0.31 |
ENST00000489715.1 |
GBE1 |
glucan (1,4-alpha-), branching enzyme 1 |
| chr1_+_10271674 | 0.31 |
ENST00000377086.1 |
KIF1B |
kinesin family member 1B |
| chr19_-_46285736 | 0.31 |
ENST00000291270.4 ENST00000447742.2 ENST00000354227.5 |
DMPK |
dystrophia myotonica-protein kinase |
| chr8_-_72274467 | 0.31 |
ENST00000340726.3 |
EYA1 |
eyes absent homolog 1 (Drosophila) |
| chr19_+_10527449 | 0.30 |
ENST00000592685.1 ENST00000380702.2 |
PDE4A |
phosphodiesterase 4A, cAMP-specific |
| chr17_+_58755184 | 0.30 |
ENST00000589222.1 ENST00000407086.3 ENST00000390652.5 |
BCAS3 |
breast carcinoma amplified sequence 3 |
| chr1_-_150849208 | 0.30 |
ENST00000358595.5 |
ARNT |
aryl hydrocarbon receptor nuclear translocator |
| chr10_+_95848824 | 0.29 |
ENST00000371385.3 ENST00000371375.1 |
PLCE1 |
phospholipase C, epsilon 1 |
| chr17_-_21156578 | 0.28 |
ENST00000399011.2 ENST00000468196.1 |
C17orf103 |
chromosome 17 open reading frame 103 |
| chr14_-_73493825 | 0.28 |
ENST00000318876.5 ENST00000556143.1 |
ZFYVE1 |
zinc finger, FYVE domain containing 1 |
| chr22_+_35695793 | 0.28 |
ENST00000456128.1 ENST00000449058.2 ENST00000411850.1 ENST00000425375.1 ENST00000436462.2 ENST00000382034.5 |
TOM1 |
target of myb1 (chicken) |
| chrX_-_150067069 | 0.28 |
ENST00000466436.1 |
CD99L2 |
CD99 molecule-like 2 |
| chr4_+_37892682 | 0.28 |
ENST00000508802.1 ENST00000261439.4 ENST00000402522.1 |
TBC1D1 |
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
| chr10_+_13142075 | 0.27 |
ENST00000378757.2 ENST00000430081.1 ENST00000378752.3 ENST00000378748.3 |
OPTN |
optineurin |
| chr2_-_152590946 | 0.27 |
ENST00000172853.10 |
NEB |
nebulin |
| chr3_+_191046810 | 0.27 |
ENST00000392455.3 ENST00000392456.3 |
CCDC50 |
coiled-coil domain containing 50 |
| chr12_+_32655048 | 0.27 |
ENST00000427716.2 ENST00000266482.3 |
FGD4 |
FYVE, RhoGEF and PH domain containing 4 |
| chr5_+_102201430 | 0.27 |
ENST00000438793.3 ENST00000346918.2 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
| chr1_-_150849174 | 0.27 |
ENST00000515192.1 |
ARNT |
aryl hydrocarbon receptor nuclear translocator |
| chr10_+_13142225 | 0.26 |
ENST00000378747.3 |
OPTN |
optineurin |
| chr11_-_67120974 | 0.26 |
ENST00000539074.1 ENST00000312419.3 |
POLD4 |
polymerase (DNA-directed), delta 4, accessory subunit |
| chr8_-_93029865 | 0.26 |
ENST00000422361.2 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr5_+_102201687 | 0.26 |
ENST00000304400.7 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
| chr11_+_6411670 | 0.26 |
ENST00000530395.1 ENST00000527275.1 |
SMPD1 |
sphingomyelin phosphodiesterase 1, acid lysosomal |
| chr14_+_23067146 | 0.26 |
ENST00000428304.2 |
ABHD4 |
abhydrolase domain containing 4 |
| chr17_-_66453562 | 0.26 |
ENST00000262139.5 ENST00000546360.1 |
WIPI1 |
WD repeat domain, phosphoinositide interacting 1 |
| chr9_+_2159850 | 0.26 |
ENST00000416751.1 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr12_-_56122426 | 0.25 |
ENST00000551173.1 |
CD63 |
CD63 molecule |
| chrX_-_101397433 | 0.25 |
ENST00000372774.3 |
TCEAL6 |
transcription elongation factor A (SII)-like 6 |
| chr1_-_182360498 | 0.25 |
ENST00000417584.2 |
GLUL |
glutamate-ammonia ligase |
| chr3_-_64211112 | 0.25 |
ENST00000295902.6 |
PRICKLE2 |
prickle homolog 2 (Drosophila) |
| chr11_+_6411636 | 0.25 |
ENST00000299397.3 ENST00000356761.2 ENST00000342245.4 |
SMPD1 |
sphingomyelin phosphodiesterase 1, acid lysosomal |
| chr5_+_149569520 | 0.25 |
ENST00000230671.2 ENST00000524041.1 |
SLC6A7 |
solute carrier family 6 (neurotransmitter transporter), member 7 |
| chr2_-_73460334 | 0.25 |
ENST00000258083.2 |
PRADC1 |
protease-associated domain containing 1 |
| chr12_-_76879852 | 0.25 |
ENST00000548341.1 |
OSBPL8 |
oxysterol binding protein-like 8 |
| chr14_-_53019211 | 0.25 |
ENST00000557374.1 ENST00000281741.4 |
TXNDC16 |
thioredoxin domain containing 16 |
| chr1_-_150669500 | 0.25 |
ENST00000271732.3 |
GOLPH3L |
golgi phosphoprotein 3-like |
| chr8_-_135522425 | 0.25 |
ENST00000521673.1 |
ZFAT |
zinc finger and AT hook domain containing |
| chr17_+_6347761 | 0.25 |
ENST00000250056.8 ENST00000571373.1 ENST00000570337.2 ENST00000572595.2 ENST00000576056.1 |
FAM64A |
family with sequence similarity 64, member A |
| chr1_-_150849047 | 0.24 |
ENST00000354396.2 ENST00000505755.1 |
ARNT |
aryl hydrocarbon receptor nuclear translocator |
| chr19_+_18699599 | 0.24 |
ENST00000450195.2 |
C19orf60 |
chromosome 19 open reading frame 60 |
| chr20_+_43160409 | 0.24 |
ENST00000372894.3 ENST00000372892.3 ENST00000372891.3 |
PKIG |
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr4_-_52904425 | 0.24 |
ENST00000535450.1 |
SGCB |
sarcoglycan, beta (43kDa dystrophin-associated glycoprotein) |
| chr20_-_18477862 | 0.23 |
ENST00000337227.4 |
RBBP9 |
retinoblastoma binding protein 9 |
| chr17_+_6347729 | 0.23 |
ENST00000572447.1 |
FAM64A |
family with sequence similarity 64, member A |
| chr10_+_94594351 | 0.23 |
ENST00000371552.4 |
EXOC6 |
exocyst complex component 6 |
| chr19_+_18699535 | 0.23 |
ENST00000358607.6 |
C19orf60 |
chromosome 19 open reading frame 60 |
| chr12_+_12938541 | 0.23 |
ENST00000356591.4 |
APOLD1 |
apolipoprotein L domain containing 1 |
| chr10_-_92681033 | 0.23 |
ENST00000371697.3 |
ANKRD1 |
ankyrin repeat domain 1 (cardiac muscle) |
| chr8_-_71519889 | 0.23 |
ENST00000521425.1 |
TRAM1 |
translocation associated membrane protein 1 |
| chr7_-_30029574 | 0.23 |
ENST00000426154.1 ENST00000421434.1 ENST00000434476.2 |
SCRN1 |
secernin 1 |
| chr4_+_86699834 | 0.23 |
ENST00000395183.2 |
ARHGAP24 |
Rho GTPase activating protein 24 |
| chr17_+_39394250 | 0.23 |
ENST00000254072.6 |
KRTAP9-8 |
keratin associated protein 9-8 |
| chr14_+_53019993 | 0.23 |
ENST00000542169.2 ENST00000555622.1 |
GPR137C |
G protein-coupled receptor 137C |
| chr22_+_30792846 | 0.23 |
ENST00000312932.9 ENST00000428195.1 |
SEC14L2 |
SEC14-like 2 (S. cerevisiae) |
| chr5_+_102201509 | 0.22 |
ENST00000348126.2 ENST00000379787.4 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
| chr17_+_45286387 | 0.22 |
ENST00000572316.1 ENST00000354968.1 ENST00000576874.1 ENST00000536623.2 |
MYL4 |
myosin, light chain 4, alkali; atrial, embryonic |
| chr17_-_42345487 | 0.22 |
ENST00000262418.6 |
SLC4A1 |
solute carrier family 4 (anion exchanger), member 1 (Diego blood group) |
| chr1_-_33647267 | 0.22 |
ENST00000291416.5 |
TRIM62 |
tripartite motif containing 62 |
| chr9_-_21305312 | 0.22 |
ENST00000259555.4 |
IFNA5 |
interferon, alpha 5 |
| chr12_+_109826524 | 0.22 |
ENST00000431443.2 |
MYO1H |
myosin IH |
| chr22_+_38071615 | 0.22 |
ENST00000215909.5 |
LGALS1 |
lectin, galactoside-binding, soluble, 1 |
| chr7_-_30029367 | 0.22 |
ENST00000242059.5 |
SCRN1 |
secernin 1 |
| chr2_+_85981008 | 0.21 |
ENST00000306279.3 |
ATOH8 |
atonal homolog 8 (Drosophila) |
| chr15_-_63448973 | 0.21 |
ENST00000462430.1 |
RPS27L |
ribosomal protein S27-like |
| chr14_+_90863327 | 0.21 |
ENST00000356978.4 |
CALM1 |
calmodulin 1 (phosphorylase kinase, delta) |
| chr7_+_18535854 | 0.21 |
ENST00000401921.1 |
HDAC9 |
histone deacetylase 9 |
| chr1_+_174844645 | 0.21 |
ENST00000486220.1 |
RABGAP1L |
RAB GTPase activating protein 1-like |
| chr5_+_102201722 | 0.21 |
ENST00000274392.9 ENST00000455264.2 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
| chr21_+_17791838 | 0.21 |
ENST00000453910.1 |
LINC00478 |
long intergenic non-protein coding RNA 478 |
| chr11_+_20044096 | 0.20 |
ENST00000533917.1 |
NAV2 |
neuron navigator 2 |
| chr17_-_79895097 | 0.20 |
ENST00000402252.2 ENST00000583564.1 ENST00000585244.1 ENST00000337943.5 ENST00000579698.1 |
PYCR1 |
pyrroline-5-carboxylate reductase 1 |
| chr2_-_65593784 | 0.20 |
ENST00000443619.2 |
SPRED2 |
sprouty-related, EVH1 domain containing 2 |
| chr17_-_1395954 | 0.20 |
ENST00000359786.5 |
MYO1C |
myosin IC |
| chr3_-_46904946 | 0.20 |
ENST00000292327.4 |
MYL3 |
myosin, light chain 3, alkali; ventricular, skeletal, slow |
| chr21_+_17791648 | 0.20 |
ENST00000602892.1 ENST00000418813.2 ENST00000435697.1 |
LINC00478 |
long intergenic non-protein coding RNA 478 |
| chr6_-_110736742 | 0.20 |
ENST00000368924.3 ENST00000368923.3 |
DDO |
D-aspartate oxidase |
| chr3_-_46904918 | 0.20 |
ENST00000395869.1 |
MYL3 |
myosin, light chain 3, alkali; ventricular, skeletal, slow |
| chr17_+_42248063 | 0.20 |
ENST00000293414.1 |
ASB16 |
ankyrin repeat and SOCS box containing 16 |
| chr12_+_104458235 | 0.20 |
ENST00000229330.4 |
HCFC2 |
host cell factor C2 |
| chr15_+_83776137 | 0.20 |
ENST00000322019.9 |
TM6SF1 |
transmembrane 6 superfamily member 1 |
| chr18_+_3448455 | 0.20 |
ENST00000549780.1 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr19_-_48018203 | 0.20 |
ENST00000595227.1 ENST00000593761.1 ENST00000263354.3 |
NAPA |
N-ethylmaleimide-sensitive factor attachment protein, alpha |
| chr1_-_182641367 | 0.20 |
ENST00000508450.1 |
RGS8 |
regulator of G-protein signaling 8 |
| chr9_-_130952989 | 0.20 |
ENST00000415526.1 ENST00000277465.4 |
CIZ1 |
CDKN1A interacting zinc finger protein 1 |
| chr8_-_74791051 | 0.19 |
ENST00000453587.2 ENST00000602969.1 ENST00000602593.1 ENST00000419880.3 ENST00000517608.1 |
UBE2W |
ubiquitin-conjugating enzyme E2W (putative) |
| chrX_+_135278908 | 0.19 |
ENST00000539015.1 ENST00000370683.1 |
FHL1 |
four and a half LIM domains 1 |
| chr12_-_123752624 | 0.19 |
ENST00000542174.1 ENST00000535796.1 |
CDK2AP1 |
cyclin-dependent kinase 2 associated protein 1 |
| chr8_+_81398444 | 0.19 |
ENST00000455036.3 ENST00000426744.2 |
ZBTB10 |
zinc finger and BTB domain containing 10 |
| chr14_+_53019822 | 0.19 |
ENST00000321662.6 |
GPR137C |
G protein-coupled receptor 137C |
| chrX_+_135279179 | 0.19 |
ENST00000370676.3 |
FHL1 |
four and a half LIM domains 1 |
| chr15_+_43809797 | 0.19 |
ENST00000399453.1 ENST00000300231.5 |
MAP1A |
microtubule-associated protein 1A |
| chr16_-_21289627 | 0.19 |
ENST00000396023.2 ENST00000415987.2 |
CRYM |
crystallin, mu |
| chr19_+_36249044 | 0.19 |
ENST00000444637.2 ENST00000396908.4 ENST00000544099.1 |
C19orf55 |
chromosome 19 open reading frame 55 |
| chr2_+_62132800 | 0.19 |
ENST00000538736.1 |
COMMD1 |
copper metabolism (Murr1) domain containing 1 |
| chr11_-_67169253 | 0.19 |
ENST00000527663.1 ENST00000312989.7 |
PPP1CA |
protein phosphatase 1, catalytic subunit, alpha isozyme |
| chr3_+_54157480 | 0.19 |
ENST00000490478.1 |
CACNA2D3 |
calcium channel, voltage-dependent, alpha 2/delta subunit 3 |
| chr1_-_19638566 | 0.18 |
ENST00000330072.5 ENST00000235835.3 |
AKR7A2 |
aldo-keto reductase family 7, member A2 (aflatoxin aldehyde reductase) |
| chr11_-_62323702 | 0.18 |
ENST00000530285.1 |
AHNAK |
AHNAK nucleoprotein |
| chr2_+_101437487 | 0.18 |
ENST00000427413.1 ENST00000542504.1 |
NPAS2 |
neuronal PAS domain protein 2 |
| chr8_+_38758737 | 0.18 |
ENST00000521746.1 ENST00000420274.1 |
PLEKHA2 |
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 2 |
| chr17_+_33570055 | 0.18 |
ENST00000299977.4 ENST00000542451.1 ENST00000592325.1 |
SLFN5 |
schlafen family member 5 |
| chr22_+_31090793 | 0.18 |
ENST00000332585.6 ENST00000382310.3 ENST00000446658.2 |
OSBP2 |
oxysterol binding protein 2 |
| chr6_+_69942298 | 0.18 |
ENST00000238918.8 |
BAI3 |
brain-specific angiogenesis inhibitor 3 |
| chr15_+_75628394 | 0.18 |
ENST00000564815.1 ENST00000338995.6 |
COMMD4 |
COMM domain containing 4 |
| chr14_-_69619823 | 0.18 |
ENST00000341516.5 |
DCAF5 |
DDB1 and CUL4 associated factor 5 |
| chr5_-_139930713 | 0.18 |
ENST00000602657.1 |
SRA1 |
steroid receptor RNA activator 1 |
| chr15_+_75628232 | 0.18 |
ENST00000267935.8 ENST00000567195.1 |
COMMD4 |
COMM domain containing 4 |
| chr17_+_39240459 | 0.18 |
ENST00000391417.4 |
KRTAP4-7 |
keratin associated protein 4-7 |
| chr17_-_79895154 | 0.18 |
ENST00000405481.4 ENST00000585215.1 ENST00000577624.1 ENST00000403172.4 |
PYCR1 |
pyrroline-5-carboxylate reductase 1 |
| chr1_-_31230650 | 0.18 |
ENST00000294507.3 |
LAPTM5 |
lysosomal protein transmembrane 5 |
| chr3_-_99833333 | 0.17 |
ENST00000354552.3 ENST00000331335.5 ENST00000398326.2 |
FILIP1L |
filamin A interacting protein 1-like |
| chr9_+_99212403 | 0.17 |
ENST00000375251.3 ENST00000375249.4 |
HABP4 |
hyaluronan binding protein 4 |
| chr17_-_79894651 | 0.17 |
ENST00000584848.1 ENST00000577756.1 ENST00000329875.8 |
PYCR1 |
pyrroline-5-carboxylate reductase 1 |
| chr10_+_22605374 | 0.17 |
ENST00000448361.1 |
COMMD3 |
COMM domain containing 3 |
| chr2_+_62132781 | 0.17 |
ENST00000311832.5 |
COMMD1 |
copper metabolism (Murr1) domain containing 1 |
| chr20_-_44485835 | 0.17 |
ENST00000457981.1 ENST00000426915.1 ENST00000217455.4 |
ACOT8 |
acyl-CoA thioesterase 8 |
| chr1_+_180165672 | 0.17 |
ENST00000443059.1 |
QSOX1 |
quiescin Q6 sulfhydryl oxidase 1 |
| chr4_-_140223670 | 0.16 |
ENST00000394228.1 ENST00000539387.1 |
NDUFC1 |
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr16_+_71929397 | 0.16 |
ENST00000537613.1 ENST00000424485.2 ENST00000606369.1 ENST00000329908.8 ENST00000538850.1 ENST00000541918.1 ENST00000534994.1 ENST00000378798.5 ENST00000539186.1 |
IST1 |
increased sodium tolerance 1 homolog (yeast) |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:1904760 | myofibroblast differentiation(GO:0036446) regulation of myofibroblast differentiation(GO:1904760) |
| 0.2 | 1.0 | GO:0018032 | protein amidation(GO:0018032) |
| 0.2 | 0.6 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.2 | 0.6 | GO:0006788 | heme oxidation(GO:0006788) smooth muscle hyperplasia(GO:0014806) negative regulation of mast cell cytokine production(GO:0032764) |
| 0.2 | 3.0 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.2 | 1.6 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.2 | 1.4 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.1 | 0.4 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.1 | 0.6 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.1 | 0.3 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.1 | 1.2 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.1 | 0.6 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 1.2 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.3 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.1 | 0.3 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.1 | 0.3 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.6 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.1 | 0.2 | GO:0061317 | canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) |
| 0.1 | 0.6 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.9 | GO:0046886 | positive regulation of hormone biosynthetic process(GO:0046886) |
| 0.1 | 0.3 | GO:1904647 | response to rotenone(GO:1904647) |
| 0.1 | 0.5 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 0.7 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 | 0.5 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.1 | 0.2 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.1 | 0.3 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 0.5 | GO:0001554 | luteolysis(GO:0001554) |
| 0.1 | 0.2 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.1 | 0.5 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.1 | 0.6 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 1.2 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 0.2 | GO:0051685 | maintenance of ER location(GO:0051685) |
| 0.1 | 0.3 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.1 | 0.2 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.1 | 0.2 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.2 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.1 | 0.2 | GO:1904404 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.1 | 0.2 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.0 | 0.6 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.3 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.2 | GO:0036496 | regulation of translational initiation by eIF2 alpha dephosphorylation(GO:0036496) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.7 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.2 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.3 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 0.0 | 0.7 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.4 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.1 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.7 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.2 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.5 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.1 | GO:0090156 | cellular sphingolipid homeostasis(GO:0090156) |
| 0.0 | 0.1 | GO:0060913 | cardiac cell fate determination(GO:0060913) |
| 0.0 | 0.1 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.0 | 0.4 | GO:0006887 | exocytosis(GO:0006887) |
| 0.0 | 0.1 | GO:0070213 | negative regulation of sister chromatid cohesion(GO:0045875) protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.6 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.3 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.1 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.0 | 0.2 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.0 | 0.2 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.2 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.0 | 0.2 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) |
| 0.0 | 0.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.3 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.7 | GO:2001046 | positive regulation of integrin-mediated signaling pathway(GO:2001046) |
| 0.0 | 0.5 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.1 | GO:0034959 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 | 0.1 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.1 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.4 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.2 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.0 | 0.3 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.1 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 | 0.5 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.3 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.1 | GO:2000334 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.3 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.1 | GO:0045348 | positive regulation of MHC class II biosynthetic process(GO:0045348) |
| 0.0 | 0.2 | GO:0038089 | positive regulation of cell migration by vascular endothelial growth factor signaling pathway(GO:0038089) |
| 0.0 | 0.1 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.1 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.0 | 0.6 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.0 | 0.1 | GO:1902954 | regulation of early endosome to recycling endosome transport(GO:1902954) |
| 0.0 | 0.3 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 0.1 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.0 | 0.6 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 0.3 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.4 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.0 | 0.1 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 2.4 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.1 | GO:2000771 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.2 | GO:0044597 | polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.0 | 0.1 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.0 | 0.5 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.0 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 0.0 | 0.2 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.2 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.1 | GO:0070253 | somatostatin secretion(GO:0070253) |
| 0.0 | 0.3 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.1 | GO:0034128 | negative regulation of MyD88-independent toll-like receptor signaling pathway(GO:0034128) |
| 0.0 | 0.1 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.0 | 0.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.2 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 1.2 | GO:0000045 | autophagosome assembly(GO:0000045) |
| 0.0 | 0.1 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.0 | 0.1 | GO:0043353 | slow-twitch skeletal muscle fiber contraction(GO:0031444) enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 1.1 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.1 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.2 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 0.0 | GO:0090675 | intermicrovillar adhesion(GO:0090675) |
| 0.0 | 0.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.0 | GO:1903336 | intralumenal vesicle formation(GO:0070676) negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.0 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.1 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.2 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.3 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.2 | GO:0051775 | response to redox state(GO:0051775) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.6 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.1 | 0.3 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 0.3 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 0.1 | 0.2 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 0.2 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.3 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.3 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.7 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.6 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.3 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.6 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.6 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.1 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.5 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.4 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.1 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 2.9 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.6 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.4 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.2 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.2 | GO:0072357 | glycogen granule(GO:0042587) PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.3 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 2.9 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.2 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.3 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.4 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.5 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.1 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.4 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 1.7 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 1.1 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.0 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.0 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.2 | 1.0 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.2 | 1.2 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.2 | 0.8 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.2 | 0.6 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.2 | 0.7 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.1 | 1.6 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.4 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.1 | 0.3 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.1 | 0.6 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 0.4 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.1 | 0.3 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.3 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.1 | 0.6 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.2 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.2 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.4 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 1.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.3 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.1 | 0.2 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.1 | 0.2 | GO:0030395 | lactose binding(GO:0030395) |
| 0.1 | 0.5 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.2 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.0 | 0.1 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.2 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.7 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 0.5 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.2 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.2 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.0 | 1.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.3 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.0 | 0.2 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.2 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.8 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.6 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.2 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 2.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.6 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.8 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.1 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.6 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.1 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.0 | 0.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.3 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.0 | GO:0015361 | low-affinity sodium:dicarboxylate symporter activity(GO:0015361) |
| 0.0 | 0.2 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.2 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.4 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.3 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 1.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.8 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.0 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.4 | GO:0001637 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 0.9 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.1 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.9 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.5 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.2 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.7 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.6 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.8 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.9 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 1.3 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.3 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |