Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
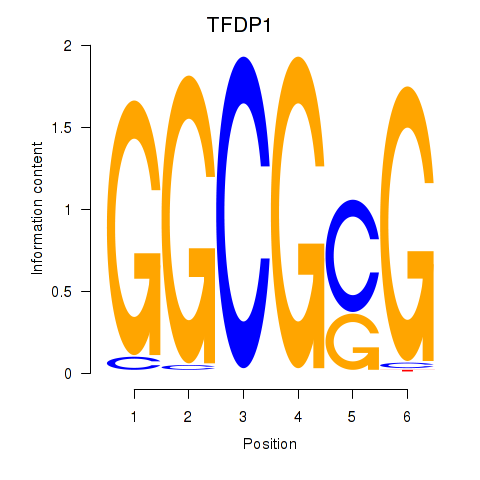
Results for TFDP1
Z-value: 1.84
Transcription factors associated with TFDP1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TFDP1
|
ENSG00000198176.8 | TFDP1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TFDP1 | hg19_v2_chr13_+_114239588_114239752 | 0.16 | 7.0e-01 | Click! |
Activity profile of TFDP1 motif
Sorted Z-values of TFDP1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TFDP1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_19263145 | 1.04 |
ENST00000532666.1 ENST00000527884.1 |
E2F8 |
E2F transcription factor 8 |
| chrX_+_30671476 | 0.96 |
ENST00000378946.3 ENST00000378943.3 ENST00000378945.3 ENST00000427190.1 ENST00000378941.3 |
GK |
glycerol kinase |
| chr6_-_86352982 | 0.70 |
ENST00000369622.3 |
SYNCRIP |
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr1_-_36235529 | 0.68 |
ENST00000318121.3 ENST00000373220.3 ENST00000520551.1 |
CLSPN |
claspin |
| chr1_-_52870104 | 0.68 |
ENST00000371568.3 |
ORC1 |
origin recognition complex, subunit 1 |
| chr12_+_122064673 | 0.67 |
ENST00000537188.1 |
ORAI1 |
ORAI calcium release-activated calcium modulator 1 |
| chr1_-_52870059 | 0.67 |
ENST00000371566.1 |
ORC1 |
origin recognition complex, subunit 1 |
| chr11_-_19262486 | 0.64 |
ENST00000250024.4 |
E2F8 |
E2F transcription factor 8 |
| chr4_+_145567173 | 0.61 |
ENST00000296575.3 |
HHIP |
hedgehog interacting protein |
| chr11_-_62389449 | 0.54 |
ENST00000534026.1 |
B3GAT3 |
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
| chr4_+_145567297 | 0.52 |
ENST00000434550.2 |
HHIP |
hedgehog interacting protein |
| chr16_-_87903079 | 0.52 |
ENST00000261622.4 |
SLC7A5 |
solute carrier family 7 (amino acid transporter light chain, L system), member 5 |
| chr2_-_40679186 | 0.52 |
ENST00000406785.2 |
SLC8A1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr4_+_94750014 | 0.50 |
ENST00000306011.3 |
ATOH1 |
atonal homolog 1 (Drosophila) |
| chr15_+_41786065 | 0.50 |
ENST00000260386.5 |
ITPKA |
inositol-trisphosphate 3-kinase A |
| chr2_+_173292390 | 0.48 |
ENST00000442250.1 ENST00000458358.1 ENST00000409080.1 |
ITGA6 |
integrin, alpha 6 |
| chr2_+_173292280 | 0.47 |
ENST00000264107.7 |
ITGA6 |
integrin, alpha 6 |
| chr2_+_36582857 | 0.47 |
ENST00000280527.2 |
CRIM1 |
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
| chr16_-_3030283 | 0.47 |
ENST00000572619.1 ENST00000574415.1 ENST00000440027.2 ENST00000572059.1 |
PKMYT1 |
protein kinase, membrane associated tyrosine/threonine 1 |
| chr3_-_48229846 | 0.45 |
ENST00000302506.3 ENST00000351231.3 ENST00000437972.1 |
CDC25A |
cell division cycle 25A |
| chr6_+_34204642 | 0.44 |
ENST00000347617.6 ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1 |
high mobility group AT-hook 1 |
| chrX_-_46618490 | 0.43 |
ENST00000328306.4 |
SLC9A7 |
solute carrier family 9, subfamily A (NHE7, cation proton antiporter 7), member 7 |
| chr2_+_173292301 | 0.43 |
ENST00000264106.6 ENST00000375221.2 ENST00000343713.4 |
ITGA6 |
integrin, alpha 6 |
| chr7_-_72936608 | 0.43 |
ENST00000404251.1 |
BAZ1B |
bromodomain adjacent to zinc finger domain, 1B |
| chr20_+_44657845 | 0.43 |
ENST00000243964.3 |
SLC12A5 |
solute carrier family 12 (potassium/chloride transporter), member 5 |
| chr14_+_65879668 | 0.42 |
ENST00000553924.1 ENST00000358307.2 ENST00000557338.1 ENST00000554610.1 |
FUT8 |
fucosyltransferase 8 (alpha (1,6) fucosyltransferase) |
| chr10_-_79398127 | 0.41 |
ENST00000372443.1 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr3_+_57994127 | 0.40 |
ENST00000490882.1 ENST00000295956.4 ENST00000358537.3 ENST00000429972.2 ENST00000348383.5 ENST00000357272.4 |
FLNB |
filamin B, beta |
| chr4_-_174255400 | 0.39 |
ENST00000506267.1 |
HMGB2 |
high mobility group box 2 |
| chr14_+_65879437 | 0.38 |
ENST00000394585.1 |
FUT8 |
fucosyltransferase 8 (alpha (1,6) fucosyltransferase) |
| chr1_+_113933371 | 0.38 |
ENST00000369617.4 |
MAGI3 |
membrane associated guanylate kinase, WW and PDZ domain containing 3 |
| chr1_+_27022839 | 0.38 |
ENST00000457599.2 |
ARID1A |
AT rich interactive domain 1A (SWI-like) |
| chr11_-_62314268 | 0.38 |
ENST00000257247.7 ENST00000531324.1 ENST00000378024.4 |
AHNAK |
AHNAK nucleoprotein |
| chr10_+_60936347 | 0.37 |
ENST00000373880.4 |
PHYHIPL |
phytanoyl-CoA 2-hydroxylase interacting protein-like |
| chr12_+_98909351 | 0.37 |
ENST00000343315.5 ENST00000266732.4 ENST00000393053.2 |
TMPO |
thymopoietin |
| chr7_-_72936531 | 0.37 |
ENST00000339594.4 |
BAZ1B |
bromodomain adjacent to zinc finger domain, 1B |
| chr1_-_226496898 | 0.37 |
ENST00000481685.1 |
LIN9 |
lin-9 homolog (C. elegans) |
| chr12_+_122459757 | 0.37 |
ENST00000261822.4 |
BCL7A |
B-cell CLL/lymphoma 7A |
| chr1_+_27022485 | 0.37 |
ENST00000324856.7 |
ARID1A |
AT rich interactive domain 1A (SWI-like) |
| chr1_+_86046433 | 0.36 |
ENST00000451137.2 |
CYR61 |
cysteine-rich, angiogenic inducer, 61 |
| chr12_+_53662110 | 0.36 |
ENST00000552462.1 |
ESPL1 |
extra spindle pole bodies homolog 1 (S. cerevisiae) |
| chr11_-_115375107 | 0.36 |
ENST00000545380.1 ENST00000452722.3 ENST00000537058.1 ENST00000536727.1 ENST00000542447.2 ENST00000331581.6 |
CADM1 |
cell adhesion molecule 1 |
| chr16_-_3030407 | 0.35 |
ENST00000431515.2 ENST00000574385.1 ENST00000576268.1 ENST00000574730.1 ENST00000575632.1 ENST00000573944.1 ENST00000262300.8 |
PKMYT1 |
protein kinase, membrane associated tyrosine/threonine 1 |
| chr17_+_34136459 | 0.35 |
ENST00000588240.1 ENST00000590273.1 ENST00000588441.1 ENST00000587272.1 ENST00000592237.1 ENST00000311979.3 |
TAF15 |
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa |
| chr8_-_67525473 | 0.35 |
ENST00000522677.3 |
MYBL1 |
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
| chr4_-_83350580 | 0.35 |
ENST00000349655.4 ENST00000602300.1 |
HNRNPDL |
heterogeneous nuclear ribonucleoprotein D-like |
| chr9_+_131218698 | 0.35 |
ENST00000434106.3 ENST00000546203.1 ENST00000446274.1 ENST00000421776.2 ENST00000432065.2 |
ODF2 |
outer dense fiber of sperm tails 2 |
| chr7_+_129251531 | 0.35 |
ENST00000393232.1 ENST00000353868.4 ENST00000539636.1 ENST00000454688.1 ENST00000223190.4 ENST00000311967.2 |
NRF1 |
nuclear respiratory factor 1 |
| chr5_-_131132658 | 0.35 |
ENST00000514667.1 ENST00000511848.1 ENST00000510461.1 |
CTC-432M15.3 FNIP1 |
Folliculin-interacting protein 1 folliculin interacting protein 1 |
| chr1_+_179923873 | 0.35 |
ENST00000367607.3 ENST00000491495.2 |
CEP350 |
centrosomal protein 350kDa |
| chr17_+_45728427 | 0.34 |
ENST00000540627.1 |
KPNB1 |
karyopherin (importin) beta 1 |
| chr9_+_131218408 | 0.34 |
ENST00000351030.3 ENST00000604420.1 ENST00000535026.1 ENST00000448249.3 ENST00000393527.3 |
ODF2 |
outer dense fiber of sperm tails 2 |
| chr4_+_84457529 | 0.34 |
ENST00000264409.4 |
AGPAT9 |
1-acylglycerol-3-phosphate O-acyltransferase 9 |
| chr20_-_32274179 | 0.34 |
ENST00000343380.5 |
E2F1 |
E2F transcription factor 1 |
| chr2_-_166651191 | 0.34 |
ENST00000392701.3 |
GALNT3 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3) |
| chr17_-_41623075 | 0.33 |
ENST00000545089.1 |
ETV4 |
ets variant 4 |
| chr8_-_124408652 | 0.33 |
ENST00000287394.5 |
ATAD2 |
ATPase family, AAA domain containing 2 |
| chr4_+_84457250 | 0.33 |
ENST00000395226.2 |
AGPAT9 |
1-acylglycerol-3-phosphate O-acyltransferase 9 |
| chr5_+_133861339 | 0.33 |
ENST00000282605.4 ENST00000361895.2 ENST00000402835.1 |
PHF15 |
jade family PHD finger 2 |
| chr12_+_98909260 | 0.32 |
ENST00000556029.1 |
TMPO |
thymopoietin |
| chr19_+_1407733 | 0.32 |
ENST00000592453.1 |
DAZAP1 |
DAZ associated protein 1 |
| chr17_+_47074758 | 0.32 |
ENST00000290341.3 |
IGF2BP1 |
insulin-like growth factor 2 mRNA binding protein 1 |
| chr1_-_54303949 | 0.32 |
ENST00000234725.8 |
NDC1 |
NDC1 transmembrane nucleoporin |
| chr16_+_12995468 | 0.32 |
ENST00000424107.3 ENST00000558583.1 ENST00000558318.1 |
SHISA9 |
shisa family member 9 |
| chr2_-_230579185 | 0.31 |
ENST00000341772.4 |
DNER |
delta/notch-like EGF repeat containing |
| chr17_-_41623009 | 0.31 |
ENST00000393664.2 |
ETV4 |
ets variant 4 |
| chr1_+_212208919 | 0.31 |
ENST00000366991.4 ENST00000542077.1 |
DTL |
denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
| chr19_+_10765003 | 0.31 |
ENST00000407004.3 ENST00000589998.1 ENST00000589600.1 |
ILF3 |
interleukin enhancer binding factor 3, 90kDa |
| chr16_-_56459354 | 0.31 |
ENST00000290649.5 |
AMFR |
autocrine motility factor receptor, E3 ubiquitin protein ligase |
| chr17_-_41623259 | 0.31 |
ENST00000538265.1 ENST00000591713.1 |
ETV4 |
ets variant 4 |
| chr15_-_73075964 | 0.31 |
ENST00000563907.1 |
ADPGK |
ADP-dependent glucokinase |
| chr14_+_101193164 | 0.31 |
ENST00000341267.4 |
DLK1 |
delta-like 1 homolog (Drosophila) |
| chr17_-_2614927 | 0.31 |
ENST00000435359.1 |
CLUH |
clustered mitochondria (cluA/CLU1) homolog |
| chr1_+_4714792 | 0.31 |
ENST00000378190.3 |
AJAP1 |
adherens junctions associated protein 1 |
| chr14_+_101193246 | 0.30 |
ENST00000331224.6 |
DLK1 |
delta-like 1 homolog (Drosophila) |
| chr17_-_38574169 | 0.30 |
ENST00000423485.1 |
TOP2A |
topoisomerase (DNA) II alpha 170kDa |
| chr12_+_53662073 | 0.30 |
ENST00000553219.1 ENST00000257934.4 |
ESPL1 |
extra spindle pole bodies homolog 1 (S. cerevisiae) |
| chr9_+_101867387 | 0.30 |
ENST00000374990.2 ENST00000552516.1 |
TGFBR1 |
transforming growth factor, beta receptor 1 |
| chr1_+_151254738 | 0.30 |
ENST00000336715.6 ENST00000324048.5 ENST00000368879.2 |
ZNF687 |
zinc finger protein 687 |
| chr2_+_42396472 | 0.29 |
ENST00000318522.5 ENST00000402711.2 |
EML4 |
echinoderm microtubule associated protein like 4 |
| chr1_-_35658736 | 0.29 |
ENST00000357214.5 |
SFPQ |
splicing factor proline/glutamine-rich |
| chr1_-_54303934 | 0.29 |
ENST00000537333.1 |
NDC1 |
NDC1 transmembrane nucleoporin |
| chr15_-_75743915 | 0.29 |
ENST00000394949.4 |
SIN3A |
SIN3 transcription regulator family member A |
| chr10_+_105127704 | 0.29 |
ENST00000369839.3 ENST00000351396.4 |
TAF5 |
TAF5 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 100kDa |
| chr10_-_106098162 | 0.29 |
ENST00000337478.1 |
ITPRIP |
inositol 1,4,5-trisphosphate receptor interacting protein |
| chr15_+_80987617 | 0.29 |
ENST00000258884.4 ENST00000558464.1 |
ABHD17C |
abhydrolase domain containing 17C |
| chr7_-_22396727 | 0.29 |
ENST00000405243.1 |
RAPGEF5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr4_-_16228083 | 0.28 |
ENST00000399920.3 |
TAPT1 |
transmembrane anterior posterior transformation 1 |
| chr7_+_150756657 | 0.28 |
ENST00000413384.2 |
SLC4A2 |
solute carrier family 4 (anion exchanger), member 2 |
| chr1_-_226496772 | 0.28 |
ENST00000359525.2 ENST00000460719.1 |
LIN9 |
lin-9 homolog (C. elegans) |
| chr3_-_52188397 | 0.28 |
ENST00000474012.1 ENST00000296484.2 |
POC1A |
POC1 centriolar protein A |
| chr18_+_43914159 | 0.28 |
ENST00000588679.1 ENST00000269439.7 ENST00000543885.1 |
RNF165 |
ring finger protein 165 |
| chr10_-_79397740 | 0.28 |
ENST00000372440.1 ENST00000480683.1 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr14_+_56585048 | 0.28 |
ENST00000267460.4 |
PELI2 |
pellino E3 ubiquitin protein ligase family member 2 |
| chr16_+_838614 | 0.28 |
ENST00000262315.9 ENST00000455171.2 |
CHTF18 |
CTF18, chromosome transmission fidelity factor 18 homolog (S. cerevisiae) |
| chr4_+_128802016 | 0.28 |
ENST00000270861.5 ENST00000515069.1 ENST00000513090.1 ENST00000507249.1 |
PLK4 |
polo-like kinase 4 |
| chr3_-_50329990 | 0.28 |
ENST00000417626.2 |
IFRD2 |
interferon-related developmental regulator 2 |
| chr4_-_57301748 | 0.28 |
ENST00000264220.2 |
PPAT |
phosphoribosyl pyrophosphate amidotransferase |
| chr3_-_120068143 | 0.27 |
ENST00000295628.3 |
LRRC58 |
leucine rich repeat containing 58 |
| chr1_+_36690011 | 0.27 |
ENST00000354618.5 ENST00000469141.2 ENST00000478853.1 |
THRAP3 |
thyroid hormone receptor associated protein 3 |
| chr2_-_208634287 | 0.27 |
ENST00000295417.3 |
FZD5 |
frizzled family receptor 5 |
| chr16_+_46723552 | 0.27 |
ENST00000219097.2 ENST00000568364.2 |
ORC6 |
origin recognition complex, subunit 6 |
| chr15_-_75744014 | 0.27 |
ENST00000394947.3 ENST00000565264.1 |
SIN3A |
SIN3 transcription regulator family member A |
| chrX_+_24167746 | 0.26 |
ENST00000428571.1 ENST00000539115.1 |
ZFX |
zinc finger protein, X-linked |
| chr8_+_27632083 | 0.26 |
ENST00000519637.1 |
ESCO2 |
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr13_-_21476900 | 0.26 |
ENST00000400602.2 ENST00000255305.6 |
XPO4 |
exportin 4 |
| chr1_-_54304212 | 0.26 |
ENST00000540001.1 |
NDC1 |
NDC1 transmembrane nucleoporin |
| chr1_-_245027766 | 0.26 |
ENST00000283179.9 |
HNRNPU |
heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) |
| chr17_-_42296855 | 0.26 |
ENST00000436088.1 |
UBTF |
upstream binding transcription factor, RNA polymerase I |
| chr8_-_67525524 | 0.26 |
ENST00000517885.1 |
MYBL1 |
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
| chr12_-_53473136 | 0.26 |
ENST00000547837.1 ENST00000301463.4 |
SPRYD3 |
SPRY domain containing 3 |
| chr17_+_29158962 | 0.26 |
ENST00000321990.4 |
ATAD5 |
ATPase family, AAA domain containing 5 |
| chr15_-_49103235 | 0.25 |
ENST00000380950.2 |
CEP152 |
centrosomal protein 152kDa |
| chr1_-_53793584 | 0.25 |
ENST00000354412.3 ENST00000347547.2 ENST00000306052.6 |
LRP8 |
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr2_+_42396574 | 0.25 |
ENST00000401738.3 |
EML4 |
echinoderm microtubule associated protein like 4 |
| chr16_+_12995614 | 0.25 |
ENST00000423335.2 |
SHISA9 |
shisa family member 9 |
| chr3_-_113465065 | 0.25 |
ENST00000497255.1 ENST00000478020.1 ENST00000240922.3 ENST00000493900.1 |
NAA50 |
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr9_-_110251836 | 0.25 |
ENST00000374672.4 |
KLF4 |
Kruppel-like factor 4 (gut) |
| chr12_-_7125770 | 0.25 |
ENST00000261407.4 |
LPCAT3 |
lysophosphatidylcholine acyltransferase 3 |
| chr17_+_37783453 | 0.25 |
ENST00000579000.1 |
PPP1R1B |
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr15_+_33022885 | 0.25 |
ENST00000322805.4 |
GREM1 |
gremlin 1, DAN family BMP antagonist |
| chrX_-_139015153 | 0.25 |
ENST00000370557.1 |
ATP11C |
ATPase, class VI, type 11C |
| chr1_-_36235559 | 0.24 |
ENST00000251195.5 |
CLSPN |
claspin |
| chr15_+_69706585 | 0.24 |
ENST00000559279.1 ENST00000395392.2 |
KIF23 |
kinesin family member 23 |
| chrX_+_64887512 | 0.24 |
ENST00000360270.5 |
MSN |
moesin |
| chr19_-_19729725 | 0.24 |
ENST00000251203.9 |
PBX4 |
pre-B-cell leukemia homeobox 4 |
| chr9_+_36572851 | 0.24 |
ENST00000298048.2 ENST00000538311.1 ENST00000536987.1 ENST00000545008.1 ENST00000536860.1 ENST00000536329.1 ENST00000541717.1 ENST00000543751.1 |
MELK |
maternal embryonic leucine zipper kinase |
| chr16_-_57831914 | 0.24 |
ENST00000421376.2 |
KIFC3 |
kinesin family member C3 |
| chr22_-_36784035 | 0.24 |
ENST00000216181.5 |
MYH9 |
myosin, heavy chain 9, non-muscle |
| chr8_+_61591337 | 0.24 |
ENST00000423902.2 |
CHD7 |
chromodomain helicase DNA binding protein 7 |
| chr17_+_47075023 | 0.24 |
ENST00000431824.2 |
IGF2BP1 |
insulin-like growth factor 2 mRNA binding protein 1 |
| chr11_+_58346584 | 0.24 |
ENST00000316059.6 |
ZFP91 |
ZFP91 zinc finger protein |
| chr15_+_99645277 | 0.24 |
ENST00000336292.6 ENST00000328642.7 |
SYNM |
synemin, intermediate filament protein |
| chr2_+_27440229 | 0.23 |
ENST00000264705.4 ENST00000403525.1 |
CAD |
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr10_-_79398250 | 0.23 |
ENST00000286627.5 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr14_-_64971288 | 0.23 |
ENST00000394715.1 |
ZBTB25 |
zinc finger and BTB domain containing 25 |
| chr6_-_86352642 | 0.23 |
ENST00000355238.6 |
SYNCRIP |
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr5_+_149109825 | 0.23 |
ENST00000360453.4 ENST00000394320.3 ENST00000309241.5 |
PPARGC1B |
peroxisome proliferator-activated receptor gamma, coactivator 1 beta |
| chr17_-_74449252 | 0.23 |
ENST00000319380.7 |
UBE2O |
ubiquitin-conjugating enzyme E2O |
| chr8_+_27632047 | 0.23 |
ENST00000397418.2 |
ESCO2 |
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr17_-_56595196 | 0.23 |
ENST00000579921.1 ENST00000579925.1 ENST00000323456.5 |
MTMR4 |
myotubularin related protein 4 |
| chr1_+_9648921 | 0.23 |
ENST00000377376.4 ENST00000340305.5 ENST00000340381.6 |
TMEM201 |
transmembrane protein 201 |
| chr17_-_13505219 | 0.23 |
ENST00000284110.1 |
HS3ST3A1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 |
| chr1_-_26185844 | 0.23 |
ENST00000538789.1 ENST00000374298.3 |
AUNIP |
aurora kinase A and ninein interacting protein |
| chr17_+_56160768 | 0.23 |
ENST00000579991.2 |
DYNLL2 |
dynein, light chain, LC8-type 2 |
| chr18_-_5296138 | 0.23 |
ENST00000400143.3 |
ZBTB14 |
zinc finger and BTB domain containing 14 |
| chr15_-_73076030 | 0.23 |
ENST00000311669.8 |
ADPGK |
ADP-dependent glucokinase |
| chr3_+_9745487 | 0.23 |
ENST00000383832.3 |
CPNE9 |
copine family member IX |
| chr8_-_95908902 | 0.23 |
ENST00000520509.1 |
CCNE2 |
cyclin E2 |
| chr6_+_43044003 | 0.23 |
ENST00000230419.4 ENST00000476760.1 ENST00000471863.1 ENST00000349241.2 ENST00000352931.2 ENST00000345201.2 |
PTK7 |
protein tyrosine kinase 7 |
| chr16_+_67063262 | 0.22 |
ENST00000565389.1 |
CBFB |
core-binding factor, beta subunit |
| chr3_+_14989076 | 0.22 |
ENST00000413118.1 ENST00000425241.1 |
NR2C2 |
nuclear receptor subfamily 2, group C, member 2 |
| chr16_+_53738053 | 0.22 |
ENST00000394647.3 |
FTO |
fat mass and obesity associated |
| chr6_+_73331776 | 0.22 |
ENST00000370398.1 |
KCNQ5 |
potassium voltage-gated channel, KQT-like subfamily, member 5 |
| chr4_+_56815102 | 0.22 |
ENST00000257287.4 |
CEP135 |
centrosomal protein 135kDa |
| chr20_+_47662805 | 0.22 |
ENST00000262982.2 ENST00000542325.1 |
CSE1L |
CSE1 chromosome segregation 1-like (yeast) |
| chr11_-_62389621 | 0.22 |
ENST00000531383.1 ENST00000265471.5 |
B3GAT3 |
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
| chr1_+_156737292 | 0.22 |
ENST00000271526.4 ENST00000353233.3 |
PRCC |
papillary renal cell carcinoma (translocation-associated) |
| chr3_-_121264848 | 0.22 |
ENST00000264233.5 |
POLQ |
polymerase (DNA directed), theta |
| chr11_+_12696102 | 0.22 |
ENST00000527636.1 ENST00000527376.1 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr7_-_111846435 | 0.22 |
ENST00000437633.1 ENST00000428084.1 |
DOCK4 |
dedicator of cytokinesis 4 |
| chr6_+_31126291 | 0.22 |
ENST00000376257.3 ENST00000376255.4 |
TCF19 |
transcription factor 19 |
| chr22_+_38201114 | 0.22 |
ENST00000340857.2 |
H1F0 |
H1 histone family, member 0 |
| chr1_-_52456352 | 0.22 |
ENST00000371655.3 |
RAB3B |
RAB3B, member RAS oncogene family |
| chr13_-_95953589 | 0.22 |
ENST00000538287.1 ENST00000376887.4 ENST00000412704.1 ENST00000536256.1 ENST00000431522.1 |
ABCC4 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 4 |
| chr4_-_149363662 | 0.22 |
ENST00000355292.3 ENST00000358102.3 |
NR3C2 |
nuclear receptor subfamily 3, group C, member 2 |
| chr1_-_6662919 | 0.22 |
ENST00000377658.4 ENST00000377663.3 |
KLHL21 |
kelch-like family member 21 |
| chr16_-_51185172 | 0.22 |
ENST00000251020.4 |
SALL1 |
spalt-like transcription factor 1 |
| chr17_-_79849438 | 0.22 |
ENST00000331204.4 ENST00000505490.2 |
ALYREF |
Aly/REF export factor |
| chrX_+_48367338 | 0.21 |
ENST00000359882.4 ENST00000537758.1 ENST00000367574.4 ENST00000355961.4 ENST00000489940.1 ENST00000361988.3 |
PORCN |
porcupine homolog (Drosophila) |
| chr1_-_159825137 | 0.21 |
ENST00000368102.1 |
C1orf204 |
chromosome 1 open reading frame 204 |
| chr9_+_130547958 | 0.21 |
ENST00000421939.1 ENST00000373265.2 |
CDK9 |
cyclin-dependent kinase 9 |
| chr11_-_64851496 | 0.21 |
ENST00000404147.3 ENST00000275517.3 |
CDCA5 |
cell division cycle associated 5 |
| chrX_+_129473859 | 0.21 |
ENST00000424447.1 |
SLC25A14 |
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr1_-_63153944 | 0.21 |
ENST00000340370.5 ENST00000404627.2 ENST00000251157.5 |
DOCK7 |
dedicator of cytokinesis 7 |
| chr6_+_31865552 | 0.21 |
ENST00000469372.1 ENST00000497706.1 |
C2 |
complement component 2 |
| chr13_-_60737898 | 0.21 |
ENST00000377908.2 ENST00000400319.1 ENST00000400320.1 ENST00000267215.4 |
DIAPH3 |
diaphanous-related formin 3 |
| chr12_+_6309517 | 0.21 |
ENST00000382519.4 ENST00000009180.4 |
CD9 |
CD9 molecule |
| chr16_+_67596310 | 0.21 |
ENST00000264010.4 ENST00000401394.1 |
CTCF |
CCCTC-binding factor (zinc finger protein) |
| chrX_+_17393543 | 0.21 |
ENST00000380060.3 |
NHS |
Nance-Horan syndrome (congenital cataracts and dental anomalies) |
| chr11_+_118307179 | 0.21 |
ENST00000534358.1 ENST00000531904.2 ENST00000389506.5 ENST00000354520.4 |
KMT2A |
lysine (K)-specific methyltransferase 2A |
| chr19_-_2456922 | 0.21 |
ENST00000582871.1 ENST00000325327.3 |
LMNB2 |
lamin B2 |
| chr3_+_110790590 | 0.21 |
ENST00000485303.1 |
PVRL3 |
poliovirus receptor-related 3 |
| chr11_-_65667884 | 0.21 |
ENST00000448083.2 ENST00000531493.1 ENST00000532401.1 |
FOSL1 |
FOS-like antigen 1 |
| chr2_+_17935119 | 0.21 |
ENST00000317402.7 |
GEN1 |
GEN1 Holliday junction 5' flap endonuclease |
| chr3_+_14989186 | 0.21 |
ENST00000435454.1 ENST00000323373.6 |
NR2C2 |
nuclear receptor subfamily 2, group C, member 2 |
| chr12_-_96794143 | 0.21 |
ENST00000543119.2 |
CDK17 |
cyclin-dependent kinase 17 |
| chr17_+_34900737 | 0.21 |
ENST00000304718.4 ENST00000485685.2 |
GGNBP2 |
gametogenetin binding protein 2 |
| chr11_-_47574664 | 0.21 |
ENST00000310513.5 ENST00000531165.1 |
CELF1 |
CUGBP, Elav-like family member 1 |
| chr12_+_66218212 | 0.21 |
ENST00000393578.3 ENST00000425208.2 ENST00000536545.1 ENST00000354636.3 |
HMGA2 |
high mobility group AT-hook 2 |
| chr1_+_89150245 | 0.21 |
ENST00000370513.5 |
PKN2 |
protein kinase N2 |
| chr18_-_70534745 | 0.21 |
ENST00000583169.1 |
NETO1 |
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr5_+_179125368 | 0.21 |
ENST00000502296.1 ENST00000504734.1 ENST00000415618.2 |
CANX |
calnexin |
| chr11_+_63706444 | 0.21 |
ENST00000377793.4 ENST00000456907.2 ENST00000539656.1 |
NAA40 |
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| chr16_+_28834303 | 0.21 |
ENST00000340394.8 ENST00000325215.6 ENST00000395547.2 ENST00000336783.4 ENST00000382686.4 ENST00000564304.1 |
ATXN2L |
ataxin 2-like |
| chr11_+_66384053 | 0.20 |
ENST00000310137.4 ENST00000393979.3 ENST00000409372.1 ENST00000443702.1 ENST00000409738.4 ENST00000514361.3 ENST00000503028.2 ENST00000412278.2 ENST00000500635.2 |
RBM14 RBM4 RBM14-RBM4 |
RNA binding motif protein 14 RNA binding motif protein 4 RBM14-RBM4 readthrough |
| chr9_-_139965000 | 0.20 |
ENST00000409687.3 |
SAPCD2 |
suppressor APC domain containing 2 |
| chr6_+_44238203 | 0.20 |
ENST00000451188.2 |
TMEM151B |
transmembrane protein 151B |
| chr4_+_54243917 | 0.20 |
ENST00000507166.1 |
FIP1L1 |
factor interacting with PAPOLA and CPSF1 |
| chr5_-_43313574 | 0.20 |
ENST00000325110.6 ENST00000433297.2 |
HMGCS1 |
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.3 | 1.0 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.3 | 0.9 | GO:0071789 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.3 | 0.8 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.2 | 0.7 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.2 | 0.3 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.2 | 1.4 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 0.7 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.1 | 0.7 | GO:0003408 | optic cup formation involved in camera-type eye development(GO:0003408) |
| 0.1 | 0.6 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.1 | 0.6 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.1 | 0.5 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.1 | 0.3 | GO:0071139 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.1 | 0.6 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.1 | 0.5 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) regulation of restriction endodeoxyribonuclease activity(GO:0032072) |
| 0.1 | 0.2 | GO:1904862 | inhibitory synapse assembly(GO:1904862) |
| 0.1 | 0.4 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.1 | 0.2 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.1 | 0.4 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.1 | 0.3 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.1 | 0.2 | GO:0043974 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.1 | 0.6 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 0.3 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.1 | 0.3 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.1 | 1.4 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.1 | 1.0 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.1 | 0.2 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.1 | 0.6 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 1.1 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.1 | 0.4 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 | 0.3 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.1 | 0.3 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.3 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.1 | 0.5 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 | 0.3 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.1 | 0.2 | GO:0032470 | positive regulation of endoplasmic reticulum calcium ion concentration(GO:0032470) |
| 0.1 | 0.2 | GO:0014740 | negative regulation of muscle hyperplasia(GO:0014740) |
| 0.1 | 0.3 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.1 | 0.4 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 0.1 | 0.2 | GO:1900158 | negative regulation of osteoclast proliferation(GO:0090291) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.1 | 0.2 | GO:1990927 | short-term synaptic potentiation(GO:1990926) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.1 | 0.4 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 0.3 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.1 | 0.2 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.1 | 1.0 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 0.2 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.1 | 0.2 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.1 | 0.2 | GO:0046832 | negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.1 | 0.2 | GO:0000967 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.1 | 0.2 | GO:0035552 | oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.1 | 0.4 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.1 | 0.4 | GO:1905075 | occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 0.1 | 0.4 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.1 | 0.4 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 0.2 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.1 | 0.2 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.1 | 0.7 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.1 | 0.1 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.1 | 0.4 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.1 | 0.3 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.1 | 0.3 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 0.2 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 0.1 | 1.0 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.1 | 0.2 | GO:0021919 | BMP signaling pathway involved in spinal cord dorsal/ventral patterning(GO:0021919) |
| 0.1 | 0.2 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) translation reinitiation(GO:0002188) |
| 0.1 | 0.2 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.1 | 0.2 | GO:1904799 | negative regulation of dendrite extension(GO:1903860) regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.1 | 0.9 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.2 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.1 | 0.4 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.1 | 0.2 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.1 | 0.7 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.2 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 | 0.2 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.1 | 0.8 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 0.6 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.1 | 0.2 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.1 | 0.4 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.1 | 0.6 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 | 0.5 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.1 | 0.2 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.1 | 0.5 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.1 | 0.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.1 | 0.2 | GO:0006679 | glucosylceramide biosynthetic process(GO:0006679) |
| 0.1 | 0.3 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.1 | 0.4 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.2 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.1 | 0.5 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.5 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.2 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.2 | GO:1900104 | hyaluranon cable assembly(GO:0036118) nephrogenic mesenchyme morphogenesis(GO:0072134) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) allantois development(GO:1905069) |
| 0.0 | 0.1 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.0 | 0.1 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 | 0.1 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 | 0.3 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.3 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.0 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.0 | 0.2 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.2 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.1 | GO:0070904 | L-ascorbic acid transport(GO:0015882) molecular hydrogen transport(GO:0015993) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.0 | 0.2 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 0.2 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 0.1 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.3 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.7 | GO:1902913 | positive regulation of neuroepithelial cell differentiation(GO:1902913) |
| 0.0 | 0.1 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.8 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.2 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.2 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.1 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.2 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.3 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 2.4 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.3 | GO:0045876 | regulation of sister chromatid cohesion(GO:0007063) positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 | 0.1 | GO:0070407 | oxidation-dependent protein catabolic process(GO:0070407) |
| 0.0 | 0.0 | GO:0046833 | positive regulation of RNA export from nucleus(GO:0046833) |
| 0.0 | 0.4 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 0.2 | GO:0061083 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.1 | GO:0036496 | regulation of translational initiation by eIF2 alpha dephosphorylation(GO:0036496) |
| 0.0 | 0.3 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.1 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) |
| 0.0 | 0.1 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.0 | 0.2 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.2 | GO:1900756 | protein processing in phagocytic vesicle(GO:1900756) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 0.0 | 0.0 | GO:1903516 | regulation of single strand break repair(GO:1903516) |
| 0.0 | 0.1 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:2000681 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.0 | 0.1 | GO:1903570 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.0 | 0.2 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.0 | 0.3 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.1 | GO:2000793 | cell proliferation involved in heart valve development(GO:2000793) |
| 0.0 | 0.1 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.0 | 0.1 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.0 | 0.4 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.4 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.4 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.2 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.0 | 0.2 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.0 | 0.2 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.0 | 0.0 | GO:1901860 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) |
| 0.0 | 0.1 | GO:1904747 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.4 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 0.2 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.0 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.0 | 0.1 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.3 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.0 | GO:0021502 | neural fold elevation formation(GO:0021502) |
| 0.0 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.0 | 0.2 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.3 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.3 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.3 | GO:0060026 | convergent extension(GO:0060026) |
| 0.0 | 0.3 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0044340 | canonical Wnt signaling pathway involved in regulation of cell proliferation(GO:0044340) |
| 0.0 | 0.1 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.6 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.2 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 1.0 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.9 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 0.1 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) |
| 0.0 | 0.3 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.0 | 0.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:0035844 | ectoderm and mesoderm interaction(GO:0007499) cloaca development(GO:0035844) cloacal septation(GO:0060197) |
| 0.0 | 0.1 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.0 | 0.1 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.3 | GO:0014870 | response to muscle inactivity(GO:0014870) |
| 0.0 | 0.1 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.0 | 0.1 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.0 | 0.1 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.2 | GO:0043634 | polyadenylation-dependent RNA catabolic process(GO:0043633) polyadenylation-dependent ncRNA catabolic process(GO:0043634) |
| 0.0 | 0.2 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.1 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.1 | GO:0060381 | regulation of single-stranded telomeric DNA binding(GO:0060380) positive regulation of single-stranded telomeric DNA binding(GO:0060381) |
| 0.0 | 0.1 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.0 | 0.4 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.1 | GO:0036333 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.0 | 0.1 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.4 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:0001172 | transcription, RNA-templated(GO:0001172) |
| 0.0 | 0.1 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) myofibroblast differentiation(GO:0036446) regulation of myofibroblast differentiation(GO:1904760) |
| 0.0 | 0.2 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.2 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.1 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.0 | 1.3 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.3 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.5 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.1 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 0.3 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.1 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.1 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 0.0 | 0.2 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.1 | GO:2000705 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.5 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.3 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.1 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.0 | 0.1 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.7 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.0 | 0.1 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.0 | 0.3 | GO:0097369 | sodium ion import(GO:0097369) |
| 0.0 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.1 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) |
| 0.0 | 0.4 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.1 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.1 | GO:0060424 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) positive regulation of branching involved in lung morphogenesis(GO:0061047) |
| 0.0 | 0.0 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.0 | 0.7 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 | 0.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.1 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.3 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) |
| 0.0 | 0.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.1 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.0 | 0.5 | GO:2000810 | regulation of bicellular tight junction assembly(GO:2000810) |
| 0.0 | 0.1 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.0 | 0.1 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.0 | 0.2 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.0 | 0.1 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.0 | 0.3 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.1 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.0 | 0.6 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 0.1 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.0 | 0.1 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.1 | GO:0035668 | TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.0 | 0.1 | GO:0006424 | glutamyl-tRNA aminoacylation(GO:0006424) |
| 0.0 | 0.1 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.1 | GO:0046108 | uridine metabolic process(GO:0046108) |
| 0.0 | 0.3 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.0 | 0.3 | GO:1903012 | positive regulation of bone development(GO:1903012) |
| 0.0 | 0.0 | GO:0002266 | follicular dendritic cell activation(GO:0002266) follicular dendritic cell differentiation(GO:0002268) |
| 0.0 | 0.1 | GO:0072711 | cellular response to hydroxyurea(GO:0072711) |
| 0.0 | 0.2 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.2 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.2 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 | 0.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.2 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.0 | 0.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.0 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.2 | GO:0051571 | positive regulation of histone H3-K4 methylation(GO:0051571) |
| 0.0 | 0.2 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.1 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.0 | 0.1 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.0 | 0.1 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.2 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.3 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.1 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.1 | GO:0048073 | regulation of eye pigmentation(GO:0048073) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.3 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.0 | 0.1 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.0 | 0.1 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.0 | 0.1 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 | 0.3 | GO:0036315 | cellular response to sterol(GO:0036315) |
| 0.0 | 0.0 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.2 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.1 | GO:0060296 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.2 | GO:0030913 | paranodal junction assembly(GO:0030913) |
| 0.0 | 0.1 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.0 | 0.2 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0070898 | RNA polymerase III transcriptional preinitiation complex assembly(GO:0070898) |
| 0.0 | 0.3 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.1 | GO:0015808 | L-alanine transport(GO:0015808) L-cystine transport(GO:0015811) |
| 0.0 | 0.1 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.0 | 0.1 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.1 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.1 | GO:0072095 | regulation of branch elongation involved in ureteric bud branching(GO:0072095) |
| 0.0 | 0.1 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.1 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.0 | 0.2 | GO:0042532 | negative regulation of tyrosine phosphorylation of STAT protein(GO:0042532) |
| 0.0 | 0.0 | GO:0042092 | type 2 immune response(GO:0042092) |
| 0.0 | 0.1 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.1 | GO:0001767 | establishment of lymphocyte polarity(GO:0001767) |
| 0.0 | 0.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.2 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.1 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.0 | GO:0021644 | vagus nerve morphogenesis(GO:0021644) chemorepulsion of branchiomotor axon(GO:0021793) |
| 0.0 | 0.2 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.1 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 | 0.1 | GO:0044854 | plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.2 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.4 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.1 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.0 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.0 | 0.1 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.1 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.0 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.2 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.5 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.0 | 0.1 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.1 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.0 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) |
| 0.0 | 0.1 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.1 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.1 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 0.4 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.1 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.0 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.0 | 0.3 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.0 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.1 | GO:0032310 | prostaglandin secretion(GO:0032310) |
| 0.0 | 0.2 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.2 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.3 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.1 | GO:0042078 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.2 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.0 | GO:0006423 | cysteinyl-tRNA aminoacylation(GO:0006423) |
| 0.0 | 0.1 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.0 | 0.0 | GO:0043506 | regulation of JUN kinase activity(GO:0043506) |
| 0.0 | 0.1 | GO:0098501 | polynucleotide dephosphorylation(GO:0098501) |
| 0.0 | 0.0 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.1 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.0 | 0.1 | GO:0042307 | positive regulation of protein import into nucleus(GO:0042307) |
| 0.0 | 0.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.4 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.0 | 0.1 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 0.1 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.0 | 0.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.0 | GO:0032849 | regulation of cellular pH reduction(GO:0032847) positive regulation of cellular pH reduction(GO:0032849) |
| 0.0 | 0.0 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.5 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.0 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.1 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.0 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.1 | GO:0061469 | response to corticotropin-releasing hormone(GO:0043435) regulation of type B pancreatic cell proliferation(GO:0061469) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.0 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.0 | 0.0 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.1 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.0 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.5 | GO:0061621 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.0 | GO:0060947 | cardiac vascular smooth muscle cell differentiation(GO:0060947) |
| 0.0 | 0.2 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.0 | 0.1 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 0.1 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 0.1 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.3 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) |
| 0.0 | 0.1 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.1 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 | 0.0 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:0043584 | nose development(GO:0043584) |
| 0.0 | 0.0 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 0.3 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.2 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.1 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.0 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.0 | GO:0051573 | negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.0 | 0.1 | GO:1904252 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.0 | 0.0 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.1 | GO:0030952 | establishment or maintenance of cytoskeleton polarity(GO:0030952) |
| 0.0 | 0.2 | GO:0046135 | pyrimidine nucleoside catabolic process(GO:0046135) |
| 0.0 | 0.0 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.0 | 0.1 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.1 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.2 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.0 | 0.1 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.1 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.0 | 0.1 | GO:0071692 | sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) |
| 0.0 | 0.0 | GO:0048936 | sensory system development(GO:0048880) peripheral nervous system neuron axonogenesis(GO:0048936) cardiac cell fate determination(GO:0060913) |
| 0.0 | 0.0 | GO:0042247 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0034676 | integrin alpha6-beta4 complex(GO:0034676) |
| 0.3 | 0.9 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.1 | 0.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 0.9 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 0.7 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 0.5 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.1 | 1.7 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.3 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.1 | 0.3 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 0.3 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.1 | 0.3 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.1 | 0.6 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.2 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.1 | 0.3 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.1 | 0.3 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.2 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 0.1 | 0.4 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.1 | 0.3 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.1 | 0.2 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.1 | 0.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 0.6 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.1 | 0.3 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.1 | 0.3 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.1 | 0.7 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 0.3 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 0.2 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.1 | 0.8 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 0.3 | GO:0001740 | Barr body(GO:0001740) |
| 0.1 | 0.2 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.1 | 0.5 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 0.4 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.1 | 1.0 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.2 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 1.5 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.2 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.4 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0071159 | NF-kappaB complex(GO:0071159) |
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.2 | GO:0031213 | RSF complex(GO:0031213) |
| 0.0 | 0.2 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.2 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.1 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 1.0 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.1 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.0 | 0.2 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.4 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.4 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.6 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.3 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.2 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.3 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.6 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.4 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.3 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.9 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.1 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.3 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.3 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.0 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.8 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.3 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.3 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.1 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.4 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.2 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 1.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 1.0 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.4 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.1 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.0 | 0.6 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.1 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.4 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.2 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.0 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.1 | GO:0016938 | kinesin I complex(GO:0016938) |
| 0.0 | 0.3 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:1990742 | microvesicle(GO:1990742) |
| 0.0 | 0.3 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 2.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.2 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.2 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.6 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.1 | GO:0043260 | laminin-10 complex(GO:0043259) laminin-11 complex(GO:0043260) |
| 0.0 | 0.2 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.2 | GO:0097346 | INO80-type complex(GO:0097346) |
| 0.0 | 0.1 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 0.1 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.1 | GO:0090576 | transcription factor TFIIIC complex(GO:0000127) RNA polymerase III transcription factor complex(GO:0090576) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.1 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.0 | 0.2 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.4 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.1 | GO:0071010 | prespliceosome(GO:0071010) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.0 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.0 | 0.1 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.0 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.0 | 0.1 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.4 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.1 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.0 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.1 | GO:0071664 | beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.0 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.2 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.2 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.0 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.2 | GO:0030666 | endocytic vesicle membrane(GO:0030666) |
| 0.0 | 1.0 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 1.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.0 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.1 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.1 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.3 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.9 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 0.1 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.2 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 1.5 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.2 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.0 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.0 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.0 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.2 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.1 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.5 | GO:0032420 | stereocilium(GO:0032420) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.3 | 0.8 | GO:0008424 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.2 | 0.8 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 0.9 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.1 | 0.8 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.1 | 0.9 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 0.5 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.1 | 1.4 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 0.3 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.1 | 0.4 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.1 | 0.3 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.1 | 0.3 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 0.1 | 0.5 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 0.2 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.1 | 0.2 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.1 | 0.2 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.1 | 0.2 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.1 | 0.3 | GO:0035501 | MH1 domain binding(GO:0035501) |
| 0.1 | 0.2 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.1 | 1.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.5 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.1 | 0.6 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.6 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.1 | 0.4 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 0.2 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.1 | 0.2 | GO:0097158 | pre-mRNA intronic pyrimidine-rich binding(GO:0097158) |
| 0.1 | 0.9 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.1 | 1.0 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 0.3 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.1 | 0.1 | GO:0004818 | glutamate-tRNA ligase activity(GO:0004818) |
| 0.1 | 0.2 | GO:0031177 | S-acetyltransferase activity(GO:0016418) phosphopantetheine binding(GO:0031177) |
| 0.1 | 0.3 | GO:0004306 | ethanolamine-phosphate cytidylyltransferase activity(GO:0004306) |
| 0.1 | 0.7 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 0.5 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.1 | 0.4 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.8 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 0.5 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.2 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.1 | 0.4 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 0.7 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.5 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 0.3 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.3 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.1 | GO:0000992 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.0 | 0.4 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.2 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.1 | GO:0008523 | L-ascorbate:sodium symporter activity(GO:0008520) sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.0 | 1.8 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 1.2 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.2 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.4 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.1 | GO:0070362 | mitochondrial light strand promoter anti-sense binding(GO:0070361) mitochondrial heavy strand promoter anti-sense binding(GO:0070362) mitochondrial heavy strand promoter sense binding(GO:0070364) |
| 0.0 | 0.2 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.4 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.4 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.4 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.2 | GO:0042835 | BRE binding(GO:0042835) |
| 0.0 | 0.2 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.1 | GO:0050405 | [hydroxymethylglutaryl-CoA reductase (NADPH)] kinase activity(GO:0047322) [acetyl-CoA carboxylase] kinase activity(GO:0050405) |
| 0.0 | 0.9 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.4 | GO:0003909 | DNA ligase activity(GO:0003909) |
| 0.0 | 1.3 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0051139 | metal ion:proton antiporter activity(GO:0051139) |
| 0.0 | 0.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.4 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.2 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 1.1 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 1.0 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.2 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.6 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.0 | 0.2 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.5 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.4 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.0 | 0.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.0 | 0.1 | GO:0005330 | dopamine:sodium symporter activity(GO:0005330) |
| 0.0 | 0.3 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.1 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.4 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.1 | GO:0032143 | single thymine insertion binding(GO:0032143) |
| 0.0 | 0.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.5 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.1 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.0 | 0.6 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.3 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.2 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.1 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.0 | 0.1 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.1 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.0 | 0.3 | GO:1901611 | phosphatidylglycerol binding(GO:1901611) |
| 0.0 | 0.1 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 1.0 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.6 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.0 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.0 | 0.1 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.0 | 0.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.2 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.2 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.5 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.7 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.2 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.4 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.1 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.0 | 0.6 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.2 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.1 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.0 | 0.9 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.2 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.2 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0001026 | TFIIIB-type transcription factor activity(GO:0001026) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.1 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) L-cystine transmembrane transporter activity(GO:0015184) alanine transmembrane transporter activity(GO:0022858) |
| 0.0 | 0.0 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.0 | 0.1 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.0 | 0.1 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.4 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.7 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.2 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.0 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.0 | 0.1 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.2 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.0 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.6 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.4 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.2 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.1 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.2 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.0 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.1 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.0 | GO:0004817 | cysteine-tRNA ligase activity(GO:0004817) |
| 0.0 | 0.1 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.0 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) |
| 0.0 | 0.1 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.2 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.1 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.0 | 0.1 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.2 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.0 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.0 | 0.2 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.0 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.0 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.0 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.0 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.0 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.0 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.6 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.3 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.2 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.0 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.0 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.1 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.2 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.2 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.2 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.0 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.0 | 0.0 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 1.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.7 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.3 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.9 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.3 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 1.0 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 2.1 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.1 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.2 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 3.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.1 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.4 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 1.3 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.0 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.7 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.6 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.4 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.0 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.0 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.1 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.2 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.1 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.1 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.3 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.1 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 0.2 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 1.8 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 0.5 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 0.1 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.1 | 0.7 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 1.2 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 1.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.1 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 1.3 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.8 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.4 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.0 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 2.1 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 0.1 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.6 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.1 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.5 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.0 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.0 | REACTOME HIV INFECTION | Genes involved in HIV Infection |
| 0.0 | 0.5 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.6 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.4 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 0.6 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.3 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.7 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.7 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.7 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 1.4 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.7 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.5 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.2 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.8 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.1 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.1 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.7 | REACTOME RNA POL II PRE TRANSCRIPTION EVENTS | Genes involved in RNA Polymerase II Pre-transcription Events |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.4 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.1 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.1 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.2 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.9 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.1 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |