Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
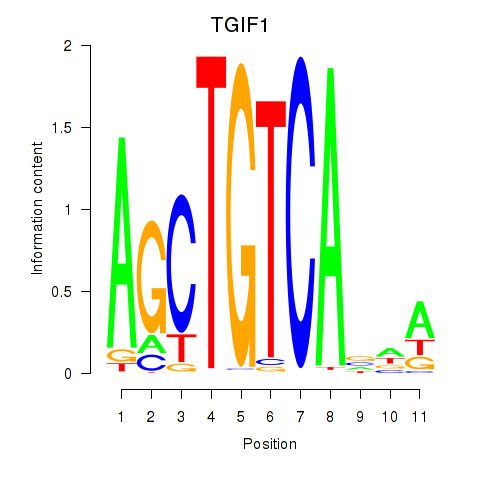
Results for TGIF1
Z-value: 1.20
Transcription factors associated with TGIF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TGIF1
|
ENSG00000177426.16 | TGIF1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TGIF1 | hg19_v2_chr18_+_3449330_3449411 | 0.53 | 1.7e-01 | Click! |
Activity profile of TGIF1 motif
Sorted Z-values of TGIF1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TGIF1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_33359646 | 2.58 |
ENST00000390003.4 ENST00000418533.2 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
| chr2_+_33359687 | 2.54 |
ENST00000402934.1 ENST00000404525.1 ENST00000407925.1 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
| chr3_+_45067659 | 2.42 |
ENST00000296130.4 |
CLEC3B |
C-type lectin domain family 3, member B |
| chr15_-_48937982 | 2.27 |
ENST00000316623.5 |
FBN1 |
fibrillin 1 |
| chr1_+_163038565 | 1.85 |
ENST00000421743.2 |
RGS4 |
regulator of G-protein signaling 4 |
| chr4_+_14113592 | 1.77 |
ENST00000502759.1 ENST00000511200.1 ENST00000512754.1 ENST00000506739.1 |
LINC01085 |
long intergenic non-protein coding RNA 1085 |
| chr11_+_6411636 | 1.35 |
ENST00000299397.3 ENST00000356761.2 ENST00000342245.4 |
SMPD1 |
sphingomyelin phosphodiesterase 1, acid lysosomal |
| chr11_+_6411670 | 1.29 |
ENST00000530395.1 ENST00000527275.1 |
SMPD1 |
sphingomyelin phosphodiesterase 1, acid lysosomal |
| chr13_-_38172863 | 1.23 |
ENST00000541481.1 ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN |
periostin, osteoblast specific factor |
| chr1_+_79086088 | 1.19 |
ENST00000370751.5 ENST00000342282.3 |
IFI44L |
interferon-induced protein 44-like |
| chr1_+_221051699 | 1.17 |
ENST00000366903.6 |
HLX |
H2.0-like homeobox |
| chr17_-_53809473 | 1.10 |
ENST00000575734.1 |
TMEM100 |
transmembrane protein 100 |
| chr2_+_24714729 | 1.06 |
ENST00000406961.1 ENST00000405141.1 |
NCOA1 |
nuclear receptor coactivator 1 |
| chr5_-_19988339 | 1.01 |
ENST00000382275.1 |
CDH18 |
cadherin 18, type 2 |
| chr4_-_175443943 | 1.00 |
ENST00000296522.6 |
HPGD |
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr4_+_55095264 | 0.95 |
ENST00000257290.5 |
PDGFRA |
platelet-derived growth factor receptor, alpha polypeptide |
| chr7_+_95401851 | 0.93 |
ENST00000447467.2 |
DYNC1I1 |
dynein, cytoplasmic 1, intermediate chain 1 |
| chr3_+_69788576 | 0.92 |
ENST00000352241.4 ENST00000448226.2 |
MITF |
microphthalmia-associated transcription factor |
| chr1_+_6615241 | 0.91 |
ENST00000333172.6 ENST00000328191.4 ENST00000351136.3 |
TAS1R1 |
taste receptor, type 1, member 1 |
| chr10_-_79398127 | 0.90 |
ENST00000372443.1 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr16_+_2880157 | 0.89 |
ENST00000382280.3 |
ZG16B |
zymogen granule protein 16B |
| chr11_+_71938925 | 0.84 |
ENST00000538751.1 |
INPPL1 |
inositol polyphosphate phosphatase-like 1 |
| chr9_-_79307096 | 0.83 |
ENST00000376717.2 ENST00000223609.6 ENST00000443509.2 |
PRUNE2 |
prune homolog 2 (Drosophila) |
| chr16_+_2880369 | 0.81 |
ENST00000572863.1 |
ZG16B |
zymogen granule protein 16B |
| chr10_-_79398250 | 0.81 |
ENST00000286627.5 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr3_+_63897605 | 0.78 |
ENST00000487717.1 |
ATXN7 |
ataxin 7 |
| chr5_+_32788945 | 0.75 |
ENST00000326958.1 |
AC026703.1 |
AC026703.1 |
| chr12_+_108908962 | 0.72 |
ENST00000552695.1 ENST00000552758.1 ENST00000361549.2 |
FICD |
FIC domain containing |
| chr1_-_149889382 | 0.71 |
ENST00000369145.1 ENST00000369146.3 |
SV2A |
synaptic vesicle glycoprotein 2A |
| chr17_-_66951474 | 0.71 |
ENST00000269080.2 |
ABCA8 |
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr2_-_145275228 | 0.69 |
ENST00000427902.1 ENST00000409487.3 ENST00000470879.1 ENST00000435831.1 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr12_+_65672423 | 0.68 |
ENST00000355192.3 ENST00000308259.5 ENST00000540804.1 ENST00000535664.1 ENST00000541189.1 |
MSRB3 |
methionine sulfoxide reductase B3 |
| chr16_+_2880254 | 0.68 |
ENST00000570670.1 |
ZG16B |
zymogen granule protein 16B |
| chr2_+_128177458 | 0.67 |
ENST00000409048.1 ENST00000422777.3 |
PROC |
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr20_+_34802295 | 0.66 |
ENST00000432603.1 |
EPB41L1 |
erythrocyte membrane protein band 4.1-like 1 |
| chr4_-_175443484 | 0.66 |
ENST00000514584.1 ENST00000542498.1 ENST00000296521.7 ENST00000422112.2 ENST00000504433.1 |
HPGD |
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr16_+_2880296 | 0.65 |
ENST00000571723.1 |
ZG16B |
zymogen granule protein 16B |
| chr5_+_141348721 | 0.65 |
ENST00000507163.1 ENST00000394519.1 |
RNF14 |
ring finger protein 14 |
| chr18_-_54305658 | 0.64 |
ENST00000586262.1 ENST00000217515.6 |
TXNL1 |
thioredoxin-like 1 |
| chr5_-_159797627 | 0.63 |
ENST00000393975.3 |
C1QTNF2 |
C1q and tumor necrosis factor related protein 2 |
| chr3_-_52719810 | 0.61 |
ENST00000424867.1 ENST00000394830.3 ENST00000431678.1 ENST00000450271.1 |
PBRM1 |
polybromo 1 |
| chr10_-_49860525 | 0.61 |
ENST00000435790.2 |
ARHGAP22 |
Rho GTPase activating protein 22 |
| chr6_+_73076432 | 0.59 |
ENST00000414192.2 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr5_+_141348640 | 0.56 |
ENST00000540015.1 ENST00000506938.1 ENST00000394514.2 ENST00000512565.1 ENST00000394515.3 |
RNF14 |
ring finger protein 14 |
| chr5_+_180257951 | 0.56 |
ENST00000501855.2 |
LINC00847 |
long intergenic non-protein coding RNA 847 |
| chrX_-_99986494 | 0.55 |
ENST00000372989.1 ENST00000455616.1 ENST00000454200.2 ENST00000276141.6 |
SYTL4 |
synaptotagmin-like 4 |
| chr3_-_127541679 | 0.55 |
ENST00000265052.5 |
MGLL |
monoglyceride lipase |
| chr5_+_180257974 | 0.54 |
ENST00000502162.2 ENST00000508309.1 ENST00000509921.1 |
LINC00847 |
long intergenic non-protein coding RNA 847 |
| chr1_-_241799232 | 0.54 |
ENST00000366553.1 |
CHML |
choroideremia-like (Rab escort protein 2) |
| chrX_-_153775426 | 0.54 |
ENST00000393562.2 |
G6PD |
glucose-6-phosphate dehydrogenase |
| chr11_+_67776012 | 0.54 |
ENST00000539229.1 |
ALDH3B1 |
aldehyde dehydrogenase 3 family, member B1 |
| chr11_+_12132117 | 0.54 |
ENST00000256194.4 |
MICAL2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
| chr12_+_59989918 | 0.53 |
ENST00000547379.1 ENST00000549465.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr11_+_46316677 | 0.53 |
ENST00000534787.1 |
CREB3L1 |
cAMP responsive element binding protein 3-like 1 |
| chr11_+_20044600 | 0.53 |
ENST00000311043.8 |
NAV2 |
neuron navigator 2 |
| chr4_-_175443788 | 0.53 |
ENST00000541923.1 |
HPGD |
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr20_-_3996165 | 0.53 |
ENST00000545616.2 ENST00000358395.6 |
RNF24 |
ring finger protein 24 |
| chr12_+_65672702 | 0.52 |
ENST00000538045.1 ENST00000535239.1 |
MSRB3 |
methionine sulfoxide reductase B3 |
| chr2_+_228678550 | 0.51 |
ENST00000409189.3 ENST00000358813.4 |
CCL20 |
chemokine (C-C motif) ligand 20 |
| chr13_+_50570019 | 0.51 |
ENST00000442421.1 |
TRIM13 |
tripartite motif containing 13 |
| chr12_-_63328817 | 0.50 |
ENST00000228705.6 |
PPM1H |
protein phosphatase, Mg2+/Mn2+ dependent, 1H |
| chr10_+_1102721 | 0.50 |
ENST00000263150.4 |
WDR37 |
WD repeat domain 37 |
| chr4_+_77870960 | 0.50 |
ENST00000505788.1 ENST00000510515.1 ENST00000504637.1 |
SEPT11 |
septin 11 |
| chr6_-_31651817 | 0.50 |
ENST00000375863.3 ENST00000375860.2 |
LY6G5C |
lymphocyte antigen 6 complex, locus G5C |
| chr12_+_59989791 | 0.49 |
ENST00000552432.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr1_-_33168336 | 0.48 |
ENST00000373484.3 |
SYNC |
syncoilin, intermediate filament protein |
| chr9_-_16728161 | 0.47 |
ENST00000603713.1 ENST00000603313.1 |
BNC2 |
basonuclin 2 |
| chr15_-_82338460 | 0.47 |
ENST00000558133.1 ENST00000329713.4 |
MEX3B |
mex-3 RNA binding family member B |
| chr4_+_166300084 | 0.46 |
ENST00000402744.4 |
CPE |
carboxypeptidase E |
| chr10_-_49812997 | 0.46 |
ENST00000417912.2 |
ARHGAP22 |
Rho GTPase activating protein 22 |
| chr10_-_14372870 | 0.46 |
ENST00000357447.2 |
FRMD4A |
FERM domain containing 4A |
| chr7_+_95401877 | 0.45 |
ENST00000524053.1 ENST00000324972.6 ENST00000537881.1 ENST00000437599.1 ENST00000359388.4 ENST00000413338.1 |
DYNC1I1 |
dynein, cytoplasmic 1, intermediate chain 1 |
| chr16_+_27413483 | 0.45 |
ENST00000337929.3 ENST00000564089.1 |
IL21R |
interleukin 21 receptor |
| chr3_-_127542051 | 0.44 |
ENST00000398104.1 |
MGLL |
monoglyceride lipase |
| chr3_-_127542021 | 0.44 |
ENST00000434178.2 |
MGLL |
monoglyceride lipase |
| chrX_-_48931648 | 0.44 |
ENST00000376386.3 ENST00000376390.4 |
PRAF2 |
PRA1 domain family, member 2 |
| chr3_-_18466026 | 0.43 |
ENST00000417717.2 |
SATB1 |
SATB homeobox 1 |
| chr3_+_106959530 | 0.42 |
ENST00000466734.1 ENST00000463143.1 ENST00000490441.1 |
LINC00883 |
long intergenic non-protein coding RNA 883 |
| chr2_-_73460334 | 0.42 |
ENST00000258083.2 |
PRADC1 |
protease-associated domain containing 1 |
| chr7_+_134551583 | 0.42 |
ENST00000435928.1 |
CALD1 |
caldesmon 1 |
| chr8_-_93029520 | 0.42 |
ENST00000521553.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr11_-_111781454 | 0.41 |
ENST00000533280.1 |
CRYAB |
crystallin, alpha B |
| chr9_+_34652164 | 0.41 |
ENST00000441545.2 ENST00000553620.1 |
IL11RA |
interleukin 11 receptor, alpha |
| chr17_-_76899275 | 0.41 |
ENST00000322630.2 ENST00000586713.1 |
DDC8 |
Protein DDC8 homolog |
| chr15_+_75498739 | 0.41 |
ENST00000565074.1 |
C15orf39 |
chromosome 15 open reading frame 39 |
| chr7_-_152133059 | 0.41 |
ENST00000262189.6 ENST00000355193.2 |
KMT2C |
lysine (K)-specific methyltransferase 2C |
| chr20_-_3996036 | 0.41 |
ENST00000336095.6 |
RNF24 |
ring finger protein 24 |
| chr17_+_68165657 | 0.40 |
ENST00000243457.3 |
KCNJ2 |
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr8_-_93029865 | 0.40 |
ENST00000422361.2 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr4_+_186990298 | 0.40 |
ENST00000296795.3 ENST00000513189.1 |
TLR3 |
toll-like receptor 3 |
| chr16_+_2588012 | 0.40 |
ENST00000354836.5 ENST00000389224.3 |
PDPK1 |
3-phosphoinositide dependent protein kinase-1 |
| chr11_-_111781554 | 0.39 |
ENST00000526167.1 ENST00000528961.1 |
CRYAB |
crystallin, alpha B |
| chr5_+_76011868 | 0.39 |
ENST00000319211.4 |
F2R |
coagulation factor II (thrombin) receptor |
| chr16_-_2260834 | 0.39 |
ENST00000562360.1 ENST00000566018.1 |
BRICD5 |
BRICHOS domain containing 5 |
| chr5_+_157170703 | 0.39 |
ENST00000286307.5 |
LSM11 |
LSM11, U7 small nuclear RNA associated |
| chr5_-_78281603 | 0.38 |
ENST00000264914.4 |
ARSB |
arylsulfatase B |
| chr5_+_140868717 | 0.38 |
ENST00000252087.1 |
PCDHGC5 |
protocadherin gamma subfamily C, 5 |
| chr9_+_17134980 | 0.38 |
ENST00000380647.3 |
CNTLN |
centlein, centrosomal protein |
| chr18_+_66465302 | 0.38 |
ENST00000360242.5 ENST00000358653.5 |
CCDC102B |
coiled-coil domain containing 102B |
| chr12_+_100041527 | 0.38 |
ENST00000324341.1 |
FAM71C |
family with sequence similarity 71, member C |
| chr10_+_124768482 | 0.37 |
ENST00000368869.4 ENST00000358776.4 |
ACADSB |
acyl-CoA dehydrogenase, short/branched chain |
| chr1_+_22379179 | 0.37 |
ENST00000315554.8 ENST00000421089.2 |
CDC42 |
cell division cycle 42 |
| chr11_-_111781610 | 0.37 |
ENST00000525823.1 |
CRYAB |
crystallin, alpha B |
| chr5_+_76012009 | 0.37 |
ENST00000505600.1 |
F2R |
coagulation factor II (thrombin) receptor |
| chr2_+_239756671 | 0.37 |
ENST00000448943.2 |
TWIST2 |
twist family bHLH transcription factor 2 |
| chr22_-_36013368 | 0.36 |
ENST00000442617.1 ENST00000397326.2 ENST00000397328.1 ENST00000451685.1 |
MB |
myoglobin |
| chr9_+_125273081 | 0.36 |
ENST00000335302.5 |
OR1J2 |
olfactory receptor, family 1, subfamily J, member 2 |
| chr12_+_55248289 | 0.36 |
ENST00000308796.6 |
MUCL1 |
mucin-like 1 |
| chr7_-_100493482 | 0.35 |
ENST00000411582.1 ENST00000419336.2 ENST00000241069.5 ENST00000302913.4 |
ACHE |
acetylcholinesterase (Yt blood group) |
| chr19_-_40791211 | 0.34 |
ENST00000579047.1 |
AKT2 |
v-akt murine thymoma viral oncogene homolog 2 |
| chr5_+_72143988 | 0.34 |
ENST00000506351.2 |
TNPO1 |
transportin 1 |
| chr16_+_50775971 | 0.34 |
ENST00000311559.9 ENST00000564326.1 ENST00000566206.1 |
CYLD |
cylindromatosis (turban tumor syndrome) |
| chr8_-_42065075 | 0.34 |
ENST00000429089.2 ENST00000519510.1 ENST00000429710.2 ENST00000524009.1 |
PLAT |
plasminogen activator, tissue |
| chr5_+_139505520 | 0.34 |
ENST00000333305.3 |
IGIP |
IgA-inducing protein |
| chr17_-_7145475 | 0.33 |
ENST00000571129.1 ENST00000571253.1 ENST00000573928.1 |
GABARAP |
GABA(A) receptor-associated protein |
| chr3_-_39234074 | 0.33 |
ENST00000340369.3 ENST00000421646.1 ENST00000396251.1 |
XIRP1 |
xin actin-binding repeat containing 1 |
| chr3_-_183735651 | 0.33 |
ENST00000427120.2 ENST00000392579.2 ENST00000382494.2 ENST00000446941.2 |
ABCC5 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr7_-_141646726 | 0.33 |
ENST00000438351.1 ENST00000439991.1 ENST00000551012.2 ENST00000546910.1 |
CLEC5A |
C-type lectin domain family 5, member A |
| chr3_-_127541194 | 0.33 |
ENST00000453507.2 |
MGLL |
monoglyceride lipase |
| chr5_-_127873896 | 0.33 |
ENST00000502468.1 |
FBN2 |
fibrillin 2 |
| chr5_-_139930713 | 0.33 |
ENST00000602657.1 |
SRA1 |
steroid receptor RNA activator 1 |
| chr4_+_128886424 | 0.32 |
ENST00000398965.1 |
C4orf29 |
chromosome 4 open reading frame 29 |
| chr6_+_108881012 | 0.32 |
ENST00000343882.6 |
FOXO3 |
forkhead box O3 |
| chr5_+_141348598 | 0.32 |
ENST00000394520.2 ENST00000347642.3 |
RNF14 |
ring finger protein 14 |
| chr16_-_67217844 | 0.31 |
ENST00000563902.1 ENST00000561621.1 ENST00000290881.7 |
KIAA0895L |
KIAA0895-like |
| chr2_+_46926048 | 0.31 |
ENST00000306503.5 |
SOCS5 |
suppressor of cytokine signaling 5 |
| chr19_-_38714847 | 0.31 |
ENST00000420980.2 ENST00000355526.4 |
DPF1 |
D4, zinc and double PHD fingers family 1 |
| chr20_-_5591626 | 0.31 |
ENST00000379019.4 |
GPCPD1 |
glycerophosphocholine phosphodiesterase GDE1 homolog (S. cerevisiae) |
| chr15_-_43559055 | 0.31 |
ENST00000220420.5 ENST00000349114.4 |
TGM5 |
transglutaminase 5 |
| chr17_+_64298944 | 0.31 |
ENST00000413366.3 |
PRKCA |
protein kinase C, alpha |
| chr9_-_99540328 | 0.31 |
ENST00000223428.4 ENST00000375231.1 ENST00000374641.3 |
ZNF510 |
zinc finger protein 510 |
| chr4_-_2420357 | 0.31 |
ENST00000511071.1 ENST00000509171.1 ENST00000290974.2 |
ZFYVE28 |
zinc finger, FYVE domain containing 28 |
| chr13_+_96329381 | 0.31 |
ENST00000602402.1 ENST00000376795.6 |
DNAJC3 |
DnaJ (Hsp40) homolog, subfamily C, member 3 |
| chr11_+_111848024 | 0.31 |
ENST00000315253.5 |
DIXDC1 |
DIX domain containing 1 |
| chr2_+_96331830 | 0.30 |
ENST00000425887.2 |
AC008268.1 |
AC008268.1 |
| chr8_-_29120580 | 0.30 |
ENST00000524189.1 |
KIF13B |
kinesin family member 13B |
| chrX_-_71525742 | 0.30 |
ENST00000450875.1 ENST00000417400.1 ENST00000431381.1 ENST00000445983.1 |
CITED1 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1 |
| chr9_-_16727978 | 0.29 |
ENST00000418777.1 ENST00000468187.2 |
BNC2 |
basonuclin 2 |
| chr15_-_37392086 | 0.29 |
ENST00000561208.1 |
MEIS2 |
Meis homeobox 2 |
| chr21_+_25801041 | 0.29 |
ENST00000453784.2 ENST00000423581.1 |
AP000476.1 |
AP000476.1 |
| chr16_+_2198604 | 0.29 |
ENST00000210187.6 |
RAB26 |
RAB26, member RAS oncogene family |
| chr11_+_120973375 | 0.29 |
ENST00000264037.2 |
TECTA |
tectorin alpha |
| chr8_+_22446763 | 0.29 |
ENST00000450780.2 ENST00000430850.2 ENST00000447849.1 |
AC037459.4 |
Uncharacterized protein |
| chr2_+_37311588 | 0.29 |
ENST00000409774.1 ENST00000608836.1 |
GPATCH11 |
G patch domain containing 11 |
| chr9_-_115653176 | 0.29 |
ENST00000374228.4 |
SLC46A2 |
solute carrier family 46, member 2 |
| chr14_+_103566481 | 0.29 |
ENST00000380069.3 |
EXOC3L4 |
exocyst complex component 3-like 4 |
| chr1_+_240177627 | 0.29 |
ENST00000447095.1 |
FMN2 |
formin 2 |
| chr3_-_190580404 | 0.29 |
ENST00000442080.1 |
GMNC |
geminin coiled-coil domain containing |
| chr8_+_71485681 | 0.28 |
ENST00000391684.1 |
AC120194.1 |
AC120194.1 |
| chr16_+_53088885 | 0.28 |
ENST00000566029.1 ENST00000447540.1 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
| chr8_+_28174649 | 0.28 |
ENST00000301908.3 |
PNOC |
prepronociceptin |
| chr12_+_15475462 | 0.28 |
ENST00000543886.1 ENST00000348962.2 |
PTPRO |
protein tyrosine phosphatase, receptor type, O |
| chr19_-_10121144 | 0.28 |
ENST00000264828.3 |
COL5A3 |
collagen, type V, alpha 3 |
| chr3_-_3221358 | 0.28 |
ENST00000424814.1 ENST00000450014.1 ENST00000231948.4 ENST00000432408.2 |
CRBN |
cereblon |
| chr17_-_7145106 | 0.28 |
ENST00000577035.1 |
GABARAP |
GABA(A) receptor-associated protein |
| chr3_+_169629354 | 0.28 |
ENST00000428432.2 ENST00000335556.3 |
SAMD7 |
sterile alpha motif domain containing 7 |
| chr6_-_111804905 | 0.28 |
ENST00000358835.3 ENST00000435970.1 |
REV3L |
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr10_+_94608245 | 0.28 |
ENST00000443748.2 ENST00000260762.6 |
EXOC6 |
exocyst complex component 6 |
| chr5_-_133968529 | 0.27 |
ENST00000402673.2 |
SAR1B |
SAR1 homolog B (S. cerevisiae) |
| chr21_+_44394620 | 0.27 |
ENST00000291547.5 |
PKNOX1 |
PBX/knotted 1 homeobox 1 |
| chr5_+_133984462 | 0.27 |
ENST00000398844.2 ENST00000322887.4 |
SEC24A |
SEC24 family member A |
| chr12_+_15475331 | 0.27 |
ENST00000281171.4 |
PTPRO |
protein tyrosine phosphatase, receptor type, O |
| chr10_-_48438974 | 0.27 |
ENST00000224605.2 |
GDF10 |
growth differentiation factor 10 |
| chr10_-_90751038 | 0.27 |
ENST00000458159.1 ENST00000415557.1 ENST00000458208.1 |
ACTA2 |
actin, alpha 2, smooth muscle, aorta |
| chr19_+_38085731 | 0.27 |
ENST00000589117.1 |
ZNF540 |
zinc finger protein 540 |
| chr11_-_111794446 | 0.27 |
ENST00000527950.1 |
CRYAB |
crystallin, alpha B |
| chr6_+_33378738 | 0.27 |
ENST00000374512.3 ENST00000374516.3 |
PHF1 |
PHD finger protein 1 |
| chr5_+_149887672 | 0.26 |
ENST00000261797.6 |
NDST1 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr16_+_50775948 | 0.26 |
ENST00000569681.1 ENST00000569418.1 ENST00000540145.1 |
CYLD |
cylindromatosis (turban tumor syndrome) |
| chr6_-_3227877 | 0.26 |
ENST00000259818.7 |
TUBB2B |
tubulin, beta 2B class IIb |
| chr17_+_36858694 | 0.26 |
ENST00000563897.1 |
CTB-58E17.1 |
CTB-58E17.1 |
| chr17_-_72358001 | 0.26 |
ENST00000375366.3 |
BTBD17 |
BTB (POZ) domain containing 17 |
| chr9_-_132805430 | 0.25 |
ENST00000446176.2 ENST00000355681.3 ENST00000420781.1 |
FNBP1 |
formin binding protein 1 |
| chrX_-_134429952 | 0.25 |
ENST00000370764.1 |
ZNF75D |
zinc finger protein 75D |
| chr3_+_112280857 | 0.25 |
ENST00000492406.1 ENST00000468642.1 |
SLC35A5 |
solute carrier family 35, member A5 |
| chr20_-_62601218 | 0.25 |
ENST00000369888.1 |
ZNF512B |
zinc finger protein 512B |
| chr5_+_134240588 | 0.24 |
ENST00000254908.6 |
PCBD2 |
pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha (TCF1) 2 |
| chr12_+_103981044 | 0.24 |
ENST00000388887.2 |
STAB2 |
stabilin 2 |
| chr5_-_132113083 | 0.24 |
ENST00000296873.7 |
SEPT8 |
septin 8 |
| chr17_+_26646175 | 0.24 |
ENST00000583381.1 ENST00000582113.1 ENST00000582384.1 |
TMEM97 |
transmembrane protein 97 |
| chr16_+_31366536 | 0.24 |
ENST00000562522.1 |
ITGAX |
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr18_+_3449330 | 0.24 |
ENST00000549253.1 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr2_+_37311645 | 0.24 |
ENST00000281932.5 |
GPATCH11 |
G patch domain containing 11 |
| chr4_-_114682364 | 0.24 |
ENST00000511664.1 |
CAMK2D |
calcium/calmodulin-dependent protein kinase II delta |
| chr1_-_53163992 | 0.24 |
ENST00000371538.3 |
SELRC1 |
cytochrome c oxidase assembly factor 7 |
| chr1_+_150039369 | 0.24 |
ENST00000369130.3 ENST00000369128.5 |
VPS45 |
vacuolar protein sorting 45 homolog (S. cerevisiae) |
| chr5_-_132112921 | 0.24 |
ENST00000378721.4 ENST00000378701.1 |
SEPT8 |
septin 8 |
| chrX_+_101967257 | 0.24 |
ENST00000543253.1 ENST00000535209.1 |
GPRASP2 |
G protein-coupled receptor associated sorting protein 2 |
| chr16_-_30582888 | 0.23 |
ENST00000563707.1 ENST00000567855.1 |
ZNF688 |
zinc finger protein 688 |
| chr14_+_31494672 | 0.23 |
ENST00000542754.2 ENST00000313566.6 |
AP4S1 |
adaptor-related protein complex 4, sigma 1 subunit |
| chr11_-_78128811 | 0.23 |
ENST00000530915.1 ENST00000361507.4 |
GAB2 |
GRB2-associated binding protein 2 |
| chr14_+_31494841 | 0.23 |
ENST00000556232.1 ENST00000216366.4 ENST00000334725.4 ENST00000554609.1 ENST00000554345.1 |
AP4S1 |
adaptor-related protein complex 4, sigma 1 subunit |
| chr15_+_75335604 | 0.23 |
ENST00000563393.1 |
PPCDC |
phosphopantothenoylcysteine decarboxylase |
| chr6_+_83777374 | 0.22 |
ENST00000349129.2 ENST00000237163.5 ENST00000536812.1 |
DOPEY1 |
dopey family member 1 |
| chr11_-_26743546 | 0.22 |
ENST00000280467.6 ENST00000396005.3 |
SLC5A12 |
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr19_+_38085768 | 0.22 |
ENST00000316433.4 ENST00000590588.1 ENST00000586134.1 ENST00000586792.1 |
ZNF540 |
zinc finger protein 540 |
| chr4_-_114682936 | 0.22 |
ENST00000454265.2 ENST00000429180.1 ENST00000418639.2 ENST00000394526.2 ENST00000296402.5 |
CAMK2D |
calcium/calmodulin-dependent protein kinase II delta |
| chr6_+_147527103 | 0.22 |
ENST00000179882.6 |
STXBP5 |
syntaxin binding protein 5 (tomosyn) |
| chr11_-_6426635 | 0.22 |
ENST00000608645.1 ENST00000608394.1 ENST00000529519.1 |
APBB1 |
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr10_+_94352956 | 0.22 |
ENST00000260731.3 |
KIF11 |
kinesin family member 11 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 7.6 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.4 | 2.6 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.4 | 1.1 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.3 | 2.4 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.3 | 1.8 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.2 | 1.0 | GO:0072275 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.2 | 1.2 | GO:0032571 | response to vitamin K(GO:0032571) bone regeneration(GO:1990523) |
| 0.2 | 2.2 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.2 | 0.7 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.2 | 1.1 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.2 | 0.7 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.2 | 0.8 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.2 | 1.5 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.2 | 0.6 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.2 | 0.5 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.2 | 0.5 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.2 | 0.5 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.1 | 0.3 | GO:1904397 | negative regulation of neuromuscular junction development(GO:1904397) |
| 0.1 | 1.7 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.1 | 0.4 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 1.0 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 1.1 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.1 | 0.5 | GO:0030070 | insulin processing(GO:0030070) |
| 0.1 | 0.3 | GO:0032223 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.1 | 1.4 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.7 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 1.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.3 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.1 | 0.3 | GO:0061110 | histone H3-T6 phosphorylation(GO:0035408) dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.1 | 0.4 | GO:0034343 | microglial cell activation involved in immune response(GO:0002282) type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.1 | 0.1 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 0.1 | 0.3 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.1 | 0.3 | GO:0009644 | response to high light intensity(GO:0009644) cellular response to light intensity(GO:0071484) |
| 0.1 | 1.4 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.7 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.1 | 0.1 | GO:0033006 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.1 | 0.2 | GO:0071338 | submandibular salivary gland formation(GO:0060661) hair follicle cell proliferation(GO:0071335) regulation of hair follicle cell proliferation(GO:0071336) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.1 | 0.2 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.1 | 0.2 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.1 | 0.8 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 0.2 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 | 0.5 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 0.2 | GO:2000566 | positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.1 | 0.5 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 0.3 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.1 | 0.5 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.1 | 0.7 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.1 | 0.3 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.1 | 0.2 | GO:0033024 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.1 | 0.2 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.1 | 0.4 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.1 | 0.2 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) beta-alanine metabolic process(GO:0019482) thymine metabolic process(GO:0019859) uracil metabolic process(GO:0019860) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.1 | 0.2 | GO:1904772 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.0 | 0.2 | GO:0036309 | protein localization to M-band(GO:0036309) |
| 0.0 | 0.4 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.4 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.4 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.0 | 0.1 | GO:0061582 | intestinal epithelial cell migration(GO:0061582) |
| 0.0 | 0.7 | GO:1901725 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) regulation of histone deacetylase activity(GO:1901725) |
| 0.0 | 0.6 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.3 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.3 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.1 | GO:0042322 | positive regulation of growth rate(GO:0040010) negative regulation of circadian sleep/wake cycle, REM sleep(GO:0042322) regulation of gastric mucosal blood circulation(GO:1904344) positive regulation of gastric mucosal blood circulation(GO:1904346) gastric mucosal blood circulation(GO:1990768) |
| 0.0 | 0.6 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.2 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.0 | 1.7 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.2 | GO:1902499 | positive regulation of protein autoubiquitination(GO:1902499) negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.6 | GO:0046321 | positive regulation of fatty acid oxidation(GO:0046321) |
| 0.0 | 0.2 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.5 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 3.2 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.0 | 1.5 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 0.8 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.3 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.6 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.2 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.4 | GO:0003012 | muscle system process(GO:0003012) |
| 0.0 | 0.1 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.0 | 0.3 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.1 | GO:0070541 | response to platinum ion(GO:0070541) |
| 0.0 | 0.2 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.1 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 0.1 | GO:0043324 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.0 | 0.1 | GO:0072229 | carbon dioxide transmembrane transport(GO:0035378) proximal convoluted tubule development(GO:0072019) metanephric proximal convoluted tubule development(GO:0072229) |
| 0.0 | 0.1 | GO:0070253 | somatostatin secretion(GO:0070253) |
| 0.0 | 0.4 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.3 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 0.3 | GO:0033033 | negative regulation of myeloid cell apoptotic process(GO:0033033) |
| 0.0 | 0.3 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.3 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.3 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.5 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.0 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.1 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.0 | 0.2 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.3 | GO:1902669 | positive regulation of axon guidance(GO:1902669) |
| 0.0 | 0.9 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.1 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 | 0.1 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.8 | GO:0003416 | endochondral bone growth(GO:0003416) |
| 0.0 | 0.4 | GO:0030101 | natural killer cell activation(GO:0030101) |
| 0.0 | 0.3 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.1 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.1 | GO:0060440 | trachea formation(GO:0060440) |
| 0.0 | 0.1 | GO:0035701 | immunoglobulin biosynthetic process(GO:0002378) hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) B cell costimulation(GO:0031296) |
| 0.0 | 0.3 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 0.8 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.2 | GO:0042262 | DNA protection(GO:0042262) |
| 0.0 | 0.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.2 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.1 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.0 | 0.1 | GO:0052250 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.0 | 0.3 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.4 | GO:0060004 | reflex(GO:0060004) |
| 0.0 | 0.7 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.0 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.2 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.0 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.0 | 0.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.5 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.5 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.9 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.8 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.1 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.2 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 | 0.3 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 0.3 | GO:0070233 | negative regulation of T cell apoptotic process(GO:0070233) |
| 0.0 | 0.4 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.0 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.0 | 0.2 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.1 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.5 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 0.3 | GO:0055070 | copper ion homeostasis(GO:0055070) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 | 0.3 | GO:0015800 | acidic amino acid transport(GO:0015800) L-glutamate transport(GO:0015813) |
| 0.0 | 0.1 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 0.2 | GO:1903817 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.1 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.4 | GO:0001652 | granular component(GO:0001652) |
| 0.3 | 7.6 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.5 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.2 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.1 | 1.4 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 1.8 | GO:0042599 | lamellar body(GO:0042599) |
| 0.1 | 0.2 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.1 | 0.5 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 0.3 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.1 | 0.2 | GO:0044753 | amphisome(GO:0044753) |
| 0.0 | 0.3 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.3 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.4 | GO:0071204 | U7 snRNP(GO:0005683) histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 1.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.5 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.3 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0020003 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 2.9 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 1.0 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.5 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.3 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 1.0 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 0.4 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.4 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.8 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.8 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0099503 | secretory vesicle(GO:0099503) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.6 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.2 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 1.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.0 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.7 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.4 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.0 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.5 | 2.2 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.3 | 2.5 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.3 | 1.0 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.2 | 1.0 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.2 | 1.7 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.2 | 0.6 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.2 | 2.6 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.2 | 1.2 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.2 | 0.5 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.2 | 0.5 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.1 | 0.4 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.1 | 0.8 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 0.3 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.1 | 0.3 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.1 | 1.8 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.4 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.1 | 0.3 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.1 | 0.2 | GO:0008124 | 4-alpha-hydroxytetrahydrobiopterin dehydratase activity(GO:0008124) |
| 0.1 | 0.7 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 1.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 0.3 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.1 | 0.3 | GO:0004306 | ethanolamine-phosphate cytidylyltransferase activity(GO:0004306) |
| 0.1 | 0.3 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.1 | 1.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 1.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 0.8 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 0.3 | GO:0042806 | fucose binding(GO:0042806) |
| 0.1 | 0.2 | GO:0052839 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.1 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.7 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.1 | 0.3 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 1.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.0 | 1.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0031768 | ghrelin receptor binding(GO:0031768) |
| 0.0 | 0.5 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.9 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.0 | 0.2 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.1 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 0.6 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.1 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.0 | 0.2 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.3 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 1.3 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.6 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.6 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.2 | GO:0030613 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.4 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.3 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.5 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.4 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.6 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.1 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 0.7 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.2 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.3 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.4 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 5.8 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.4 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.2 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.0 | 0.1 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.3 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.7 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.2 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.0 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) indanol dehydrogenase activity(GO:0047718) |
| 0.0 | 0.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.2 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.0 | GO:0000384 | first spliceosomal transesterification activity(GO:0000384) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 2.9 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 1.1 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.0 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.0 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 2.0 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 6.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.6 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.3 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.7 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.5 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 3.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.5 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.5 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.7 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.1 | 1.8 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 1.0 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 2.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.7 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.6 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.9 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 1.1 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.6 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.8 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 1.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.6 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 2.0 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.9 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.6 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 1.1 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.3 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.4 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.4 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 1.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.7 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.4 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |