Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
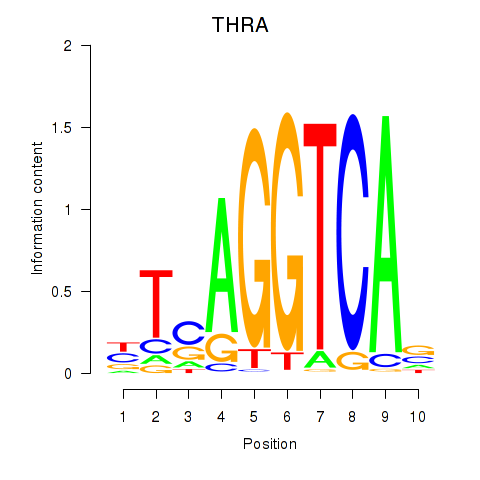
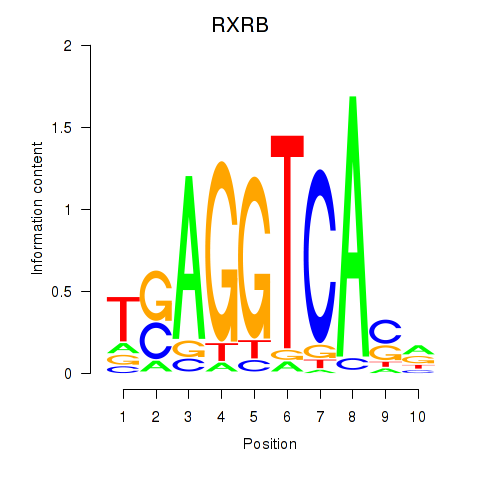
Results for THRA_RXRB
Z-value: 0.88
Transcription factors associated with THRA_RXRB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
THRA
|
ENSG00000126351.8 | THRA |
|
RXRB
|
ENSG00000204231.6 | RXRB |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| THRA | hg19_v2_chr17_+_38219063_38219154 | -0.64 | 8.7e-02 | Click! |
| RXRB | hg19_v2_chr6_-_33168391_33168465 | -0.46 | 2.5e-01 | Click! |
Activity profile of THRA_RXRB motif
Sorted Z-values of THRA_RXRB motif
Network of associatons between targets according to the STRING database.
First level regulatory network of THRA_RXRB
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_242612779 | 2.24 |
ENST00000427495.1 |
PLD5 |
phospholipase D family, member 5 |
| chr1_-_175162048 | 1.44 |
ENST00000444639.1 |
KIAA0040 |
KIAA0040 |
| chr15_+_45422178 | 1.32 |
ENST00000389037.3 ENST00000558322.1 |
DUOX1 |
dual oxidase 1 |
| chr15_+_45422131 | 1.31 |
ENST00000321429.4 |
DUOX1 |
dual oxidase 1 |
| chr1_-_175161890 | 1.30 |
ENST00000545251.2 ENST00000423313.1 |
KIAA0040 |
KIAA0040 |
| chr14_+_67999999 | 1.14 |
ENST00000329153.5 |
PLEKHH1 |
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1 |
| chr2_+_220491973 | 1.09 |
ENST00000358055.3 |
SLC4A3 |
solute carrier family 4 (anion exchanger), member 3 |
| chr19_-_49565254 | 1.07 |
ENST00000593537.1 |
NTF4 |
neurotrophin 4 |
| chr2_+_220492116 | 0.83 |
ENST00000373760.2 |
SLC4A3 |
solute carrier family 4 (anion exchanger), member 3 |
| chr6_+_125524785 | 0.76 |
ENST00000392482.2 |
TPD52L1 |
tumor protein D52-like 1 |
| chr3_-_190167571 | 0.74 |
ENST00000354905.2 |
TMEM207 |
transmembrane protein 207 |
| chr12_-_12715266 | 0.72 |
ENST00000228862.2 |
DUSP16 |
dual specificity phosphatase 16 |
| chr1_+_152881014 | 0.69 |
ENST00000368764.3 ENST00000392667.2 |
IVL |
involucrin |
| chr22_-_20255212 | 0.67 |
ENST00000416372.1 |
RTN4R |
reticulon 4 receptor |
| chr19_+_45281118 | 0.63 |
ENST00000270279.3 ENST00000341505.4 |
CBLC |
Cbl proto-oncogene C, E3 ubiquitin protein ligase |
| chr18_+_19749386 | 0.61 |
ENST00000269216.3 |
GATA6 |
GATA binding protein 6 |
| chr7_+_73245193 | 0.58 |
ENST00000340958.2 |
CLDN4 |
claudin 4 |
| chr9_-_104249319 | 0.56 |
ENST00000374847.1 |
TMEM246 |
transmembrane protein 246 |
| chr19_+_54371114 | 0.55 |
ENST00000448420.1 ENST00000439000.1 ENST00000391770.4 ENST00000391771.1 |
MYADM |
myeloid-associated differentiation marker |
| chr12_-_95510743 | 0.55 |
ENST00000551521.1 |
FGD6 |
FYVE, RhoGEF and PH domain containing 6 |
| chr15_+_40531621 | 0.54 |
ENST00000560346.1 |
PAK6 |
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr15_-_75017711 | 0.54 |
ENST00000567032.1 ENST00000564596.1 ENST00000566503.1 ENST00000395049.4 ENST00000395048.2 ENST00000379727.3 |
CYP1A1 |
cytochrome P450, family 1, subfamily A, polypeptide 1 |
| chr14_-_55369525 | 0.53 |
ENST00000543643.2 ENST00000536224.2 ENST00000395514.1 ENST00000491895.2 |
GCH1 |
GTP cyclohydrolase 1 |
| chr8_-_144651024 | 0.52 |
ENST00000524906.1 ENST00000532862.1 ENST00000534459.1 |
MROH6 |
maestro heat-like repeat family member 6 |
| chr3_+_14989186 | 0.52 |
ENST00000435454.1 ENST00000323373.6 |
NR2C2 |
nuclear receptor subfamily 2, group C, member 2 |
| chr11_-_119599794 | 0.50 |
ENST00000264025.3 |
PVRL1 |
poliovirus receptor-related 1 (herpesvirus entry mediator C) |
| chrX_-_152989798 | 0.49 |
ENST00000441714.1 ENST00000442093.1 ENST00000429550.1 ENST00000345046.6 |
BCAP31 |
B-cell receptor-associated protein 31 |
| chr4_+_20255123 | 0.48 |
ENST00000504154.1 ENST00000273739.5 |
SLIT2 |
slit homolog 2 (Drosophila) |
| chr6_-_3457256 | 0.48 |
ENST00000436008.2 |
SLC22A23 |
solute carrier family 22, member 23 |
| chr16_+_85942594 | 0.46 |
ENST00000566369.1 |
IRF8 |
interferon regulatory factor 8 |
| chr22_-_37584321 | 0.45 |
ENST00000397110.2 ENST00000337843.2 |
C1QTNF6 |
C1q and tumor necrosis factor related protein 6 |
| chr2_+_230787201 | 0.45 |
ENST00000283946.3 |
FBXO36 |
F-box protein 36 |
| chr5_+_66124590 | 0.45 |
ENST00000490016.2 ENST00000403666.1 ENST00000450827.1 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
| chr11_+_1860200 | 0.44 |
ENST00000381911.1 |
TNNI2 |
troponin I type 2 (skeletal, fast) |
| chr10_+_71389983 | 0.43 |
ENST00000373279.4 |
C10orf35 |
chromosome 10 open reading frame 35 |
| chr2_+_230787213 | 0.43 |
ENST00000409992.1 |
FBXO36 |
F-box protein 36 |
| chr22_-_29137771 | 0.43 |
ENST00000439200.1 ENST00000405598.1 ENST00000398017.2 ENST00000425190.2 ENST00000348295.3 ENST00000382578.1 ENST00000382565.1 ENST00000382566.1 ENST00000382580.2 ENST00000328354.6 |
CHEK2 |
checkpoint kinase 2 |
| chr2_-_74779744 | 0.40 |
ENST00000409249.1 |
LOXL3 |
lysyl oxidase-like 3 |
| chr1_+_160147176 | 0.39 |
ENST00000470705.1 |
ATP1A4 |
ATPase, Na+/K+ transporting, alpha 4 polypeptide |
| chr12_+_41221794 | 0.38 |
ENST00000547849.1 |
CNTN1 |
contactin 1 |
| chr17_-_27503770 | 0.38 |
ENST00000533112.1 |
MYO18A |
myosin XVIIIA |
| chr10_-_116164450 | 0.38 |
ENST00000369271.3 |
AFAP1L2 |
actin filament associated protein 1-like 2 |
| chr10_-_116164239 | 0.37 |
ENST00000419268.1 ENST00000304129.4 ENST00000545353.1 |
AFAP1L2 |
actin filament associated protein 1-like 2 |
| chr5_+_162887556 | 0.37 |
ENST00000393915.4 ENST00000432118.2 ENST00000358715.3 |
HMMR |
hyaluronan-mediated motility receptor (RHAMM) |
| chr19_-_52227221 | 0.35 |
ENST00000222115.1 ENST00000540069.2 |
HAS1 |
hyaluronan synthase 1 |
| chr19_-_6433765 | 0.34 |
ENST00000321510.6 |
SLC25A41 |
solute carrier family 25, member 41 |
| chr6_-_99797522 | 0.33 |
ENST00000389677.5 |
FAXC |
failed axon connections homolog (Drosophila) |
| chr17_-_4458616 | 0.33 |
ENST00000381556.2 |
MYBBP1A |
MYB binding protein (P160) 1a |
| chrX_-_117250740 | 0.32 |
ENST00000371882.1 ENST00000540167.1 ENST00000545703.1 |
KLHL13 |
kelch-like family member 13 |
| chr2_-_165698521 | 0.31 |
ENST00000409184.3 ENST00000392717.2 ENST00000456693.1 |
COBLL1 |
cordon-bleu WH2 repeat protein-like 1 |
| chr11_+_1856034 | 0.30 |
ENST00000341958.3 |
SYT8 |
synaptotagmin VIII |
| chr15_-_45422056 | 0.30 |
ENST00000267803.4 ENST00000559014.1 ENST00000558851.1 ENST00000559988.1 ENST00000558996.1 ENST00000558422.1 ENST00000559226.1 ENST00000558326.1 ENST00000558377.1 ENST00000559644.1 |
DUOXA1 |
dual oxidase maturation factor 1 |
| chr8_+_103563792 | 0.29 |
ENST00000285402.3 |
ODF1 |
outer dense fiber of sperm tails 1 |
| chr11_-_123756334 | 0.29 |
ENST00000528595.1 ENST00000375026.2 |
TMEM225 |
transmembrane protein 225 |
| chr15_-_34629922 | 0.29 |
ENST00000559484.1 ENST00000354181.3 ENST00000558589.1 ENST00000458406.2 |
SLC12A6 |
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr8_+_110552831 | 0.29 |
ENST00000530629.1 |
EBAG9 |
estrogen receptor binding site associated, antigen, 9 |
| chr3_+_10068095 | 0.28 |
ENST00000287647.3 ENST00000383807.1 ENST00000383806.1 ENST00000419585.1 |
FANCD2 |
Fanconi anemia, complementation group D2 |
| chr11_+_1855645 | 0.28 |
ENST00000381968.3 ENST00000381978.3 |
SYT8 |
synaptotagmin VIII |
| chr20_+_1875110 | 0.28 |
ENST00000400068.3 |
SIRPA |
signal-regulatory protein alpha |
| chr20_+_1875378 | 0.28 |
ENST00000356025.3 |
SIRPA |
signal-regulatory protein alpha |
| chr6_+_31783291 | 0.28 |
ENST00000375651.5 ENST00000608703.1 ENST00000458062.2 |
HSPA1A |
heat shock 70kDa protein 1A |
| chr17_+_37894570 | 0.27 |
ENST00000394211.3 |
GRB7 |
growth factor receptor-bound protein 7 |
| chr19_+_39989580 | 0.26 |
ENST00000596614.1 ENST00000205143.4 |
DLL3 |
delta-like 3 (Drosophila) |
| chr15_-_34630234 | 0.26 |
ENST00000558667.1 ENST00000561120.1 ENST00000559236.1 ENST00000397702.2 |
SLC12A6 |
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr2_-_165698662 | 0.26 |
ENST00000194871.6 ENST00000445474.2 |
COBLL1 |
cordon-bleu WH2 repeat protein-like 1 |
| chr4_+_177241094 | 0.26 |
ENST00000503362.1 |
SPCS3 |
signal peptidase complex subunit 3 homolog (S. cerevisiae) |
| chr14_+_22293618 | 0.25 |
ENST00000390432.2 |
TRAV10 |
T cell receptor alpha variable 10 |
| chr10_+_77056134 | 0.25 |
ENST00000528121.1 ENST00000416398.1 |
ZNF503-AS1 |
ZNF503 antisense RNA 1 |
| chr2_-_74667612 | 0.25 |
ENST00000305557.5 ENST00000233330.6 |
RTKN |
rhotekin |
| chr14_-_102552659 | 0.25 |
ENST00000441629.2 |
HSP90AA1 |
heat shock protein 90kDa alpha (cytosolic), class A member 1 |
| chr16_-_72206034 | 0.24 |
ENST00000537465.1 ENST00000237353.10 |
PMFBP1 |
polyamine modulated factor 1 binding protein 1 |
| chr12_-_54779511 | 0.24 |
ENST00000551109.1 ENST00000546970.1 |
ZNF385A |
zinc finger protein 385A |
| chr17_+_6899366 | 0.24 |
ENST00000251535.6 |
ALOX12 |
arachidonate 12-lipoxygenase |
| chr22_-_29784519 | 0.24 |
ENST00000357586.2 ENST00000356015.2 ENST00000432560.2 ENST00000317368.7 |
AP1B1 |
adaptor-related protein complex 1, beta 1 subunit |
| chr10_+_26986582 | 0.24 |
ENST00000376215.5 ENST00000376203.5 |
PDSS1 |
prenyl (decaprenyl) diphosphate synthase, subunit 1 |
| chr17_+_48046538 | 0.24 |
ENST00000240306.3 |
DLX4 |
distal-less homeobox 4 |
| chr7_-_151433393 | 0.23 |
ENST00000492843.1 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr17_+_37894179 | 0.23 |
ENST00000577695.1 ENST00000309156.4 ENST00000309185.3 |
GRB7 |
growth factor receptor-bound protein 7 |
| chr7_-_151433342 | 0.23 |
ENST00000433631.2 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr17_-_39216344 | 0.23 |
ENST00000391418.2 |
KRTAP2-3 |
keratin associated protein 2-3 |
| chr11_-_58343319 | 0.23 |
ENST00000395074.2 |
LPXN |
leupaxin |
| chr19_+_1065922 | 0.22 |
ENST00000539243.2 |
HMHA1 |
histocompatibility (minor) HA-1 |
| chr1_-_204329013 | 0.22 |
ENST00000272203.3 ENST00000414478.1 |
PLEKHA6 |
pleckstrin homology domain containing, family A member 6 |
| chr14_-_24036943 | 0.21 |
ENST00000556843.1 ENST00000397120.3 ENST00000557189.1 |
AP1G2 |
adaptor-related protein complex 1, gamma 2 subunit |
| chr17_-_41132088 | 0.21 |
ENST00000591916.1 ENST00000451885.2 ENST00000454303.1 |
PTGES3L PTGES3L-AARSD1 |
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr6_+_41196077 | 0.21 |
ENST00000448827.2 |
TREML4 |
triggering receptor expressed on myeloid cells-like 4 |
| chr6_+_41196052 | 0.21 |
ENST00000341495.2 |
TREML4 |
triggering receptor expressed on myeloid cells-like 4 |
| chr10_+_81107271 | 0.20 |
ENST00000448165.1 |
PPIF |
peptidylprolyl isomerase F |
| chr22_-_36784035 | 0.20 |
ENST00000216181.5 |
MYH9 |
myosin, heavy chain 9, non-muscle |
| chr2_+_74757050 | 0.20 |
ENST00000352222.3 ENST00000437202.1 |
HTRA2 |
HtrA serine peptidase 2 |
| chr1_-_33366931 | 0.20 |
ENST00000373463.3 ENST00000329151.5 |
TMEM54 |
transmembrane protein 54 |
| chr22_+_51176624 | 0.20 |
ENST00000216139.5 ENST00000529621.1 |
ACR |
acrosin |
| chr2_+_65663812 | 0.19 |
ENST00000606978.1 ENST00000377977.3 ENST00000536804.1 |
AC074391.1 |
AC074391.1 |
| chr3_-_50340996 | 0.19 |
ENST00000266031.4 ENST00000395143.2 ENST00000457214.2 ENST00000447605.2 ENST00000418723.1 ENST00000395144.2 |
HYAL1 |
hyaluronoglucosaminidase 1 |
| chr19_+_39989535 | 0.19 |
ENST00000356433.5 |
DLL3 |
delta-like 3 (Drosophila) |
| chr13_-_46756351 | 0.19 |
ENST00000323076.2 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
| chr17_-_1928621 | 0.19 |
ENST00000331238.6 |
RTN4RL1 |
reticulon 4 receptor-like 1 |
| chr11_+_55594695 | 0.19 |
ENST00000378397.1 |
OR5L2 |
olfactory receptor, family 5, subfamily L, member 2 |
| chr1_+_70876926 | 0.18 |
ENST00000370938.3 ENST00000346806.2 |
CTH |
cystathionase (cystathionine gamma-lyase) |
| chr11_-_59578202 | 0.18 |
ENST00000300151.4 |
MRPL16 |
mitochondrial ribosomal protein L16 |
| chr6_+_35773070 | 0.18 |
ENST00000373853.1 ENST00000360215.1 |
LHFPL5 |
lipoma HMGIC fusion partner-like 5 |
| chr15_-_41408409 | 0.18 |
ENST00000361937.3 |
INO80 |
INO80 complex subunit |
| chr22_-_29138386 | 0.18 |
ENST00000544772.1 |
CHEK2 |
checkpoint kinase 2 |
| chr9_+_27109392 | 0.18 |
ENST00000406359.4 |
TEK |
TEK tyrosine kinase, endothelial |
| chr7_-_105319536 | 0.18 |
ENST00000477775.1 |
ATXN7L1 |
ataxin 7-like 1 |
| chr11_+_66624527 | 0.17 |
ENST00000393952.3 |
LRFN4 |
leucine rich repeat and fibronectin type III domain containing 4 |
| chr1_-_205904950 | 0.17 |
ENST00000340781.4 |
SLC26A9 |
solute carrier family 26 (anion exchanger), member 9 |
| chr15_-_44487408 | 0.17 |
ENST00000402883.1 ENST00000417257.1 |
FRMD5 |
FERM domain containing 5 |
| chr11_-_62752162 | 0.17 |
ENST00000458333.2 ENST00000421062.2 |
SLC22A6 |
solute carrier family 22 (organic anion transporter), member 6 |
| chr1_+_70876891 | 0.17 |
ENST00000411986.2 |
CTH |
cystathionase (cystathionine gamma-lyase) |
| chr6_+_30614779 | 0.17 |
ENST00000293604.6 ENST00000376473.5 |
C6orf136 |
chromosome 6 open reading frame 136 |
| chr11_+_33563821 | 0.17 |
ENST00000321505.4 ENST00000265654.5 ENST00000389726.3 |
KIAA1549L |
KIAA1549-like |
| chr2_+_173686303 | 0.17 |
ENST00000397087.3 |
RAPGEF4 |
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr12_+_102514019 | 0.17 |
ENST00000537257.1 ENST00000358383.5 ENST00000392911.2 |
PARPBP |
PARP1 binding protein |
| chr17_-_18161870 | 0.16 |
ENST00000579294.1 ENST00000545457.2 ENST00000379450.4 ENST00000578558.1 |
FLII |
flightless I homolog (Drosophila) |
| chr16_-_50715196 | 0.16 |
ENST00000423026.2 |
SNX20 |
sorting nexin 20 |
| chr16_-_4466565 | 0.16 |
ENST00000572467.1 ENST00000423908.2 ENST00000572044.1 ENST00000571052.1 |
CORO7-PAM16 CORO7 |
CORO7-PAM16 readthrough coronin 7 |
| chr19_-_49176264 | 0.16 |
ENST00000270235.4 ENST00000596844.1 |
NTN5 |
netrin 5 |
| chr2_-_230786619 | 0.16 |
ENST00000389045.3 ENST00000409677.1 |
TRIP12 |
thyroid hormone receptor interactor 12 |
| chr13_+_76123883 | 0.15 |
ENST00000377595.3 |
UCHL3 |
ubiquitin carboxyl-terminal esterase L3 (ubiquitin thiolesterase) |
| chrX_-_132887729 | 0.15 |
ENST00000406757.2 |
GPC3 |
glypican 3 |
| chr13_+_113030625 | 0.15 |
ENST00000283550.3 |
SPACA7 |
sperm acrosome associated 7 |
| chr8_+_63161491 | 0.15 |
ENST00000523211.1 ENST00000524201.1 |
NKAIN3 |
Na+/K+ transporting ATPase interacting 3 |
| chr14_-_103523745 | 0.15 |
ENST00000361246.2 |
CDC42BPB |
CDC42 binding protein kinase beta (DMPK-like) |
| chr4_+_7194247 | 0.15 |
ENST00000507866.2 |
SORCS2 |
sortilin-related VPS10 domain containing receptor 2 |
| chr11_-_77122928 | 0.15 |
ENST00000528203.1 ENST00000528592.1 ENST00000528633.1 ENST00000529248.1 |
PAK1 |
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr6_+_30615167 | 0.15 |
ENST00000446773.2 |
C6orf136 |
chromosome 6 open reading frame 136 |
| chr12_+_57984965 | 0.15 |
ENST00000540759.2 ENST00000551772.1 ENST00000550465.1 ENST00000354947.5 |
PIP4K2C |
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma |
| chr12_+_102513950 | 0.15 |
ENST00000378128.3 ENST00000327680.2 ENST00000541394.1 ENST00000543784.1 |
PARPBP |
PARP1 binding protein |
| chr6_+_30614886 | 0.15 |
ENST00000376471.4 |
C6orf136 |
chromosome 6 open reading frame 136 |
| chr12_-_102513843 | 0.15 |
ENST00000551744.2 ENST00000552283.1 |
NUP37 |
nucleoporin 37kDa |
| chr7_+_142982023 | 0.14 |
ENST00000359333.3 ENST00000409244.1 ENST00000409541.1 ENST00000410004.1 |
TMEM139 |
transmembrane protein 139 |
| chr3_-_98241760 | 0.14 |
ENST00000507874.1 ENST00000502299.1 ENST00000508659.1 ENST00000510545.1 ENST00000511667.1 ENST00000394185.2 ENST00000394181.2 ENST00000508902.1 ENST00000341181.6 ENST00000437922.1 ENST00000394180.2 |
CLDND1 |
claudin domain containing 1 |
| chr5_-_176056974 | 0.14 |
ENST00000510387.1 ENST00000506696.1 |
SNCB |
synuclein, beta |
| chr22_+_29138013 | 0.14 |
ENST00000216027.3 ENST00000398941.2 |
HSCB |
HscB mitochondrial iron-sulfur cluster co-chaperone |
| chr14_-_106054659 | 0.14 |
ENST00000390539.2 |
IGHA2 |
immunoglobulin heavy constant alpha 2 (A2m marker) |
| chr22_+_23243156 | 0.14 |
ENST00000390323.2 |
IGLC2 |
immunoglobulin lambda constant 2 (Kern-Oz- marker) |
| chr8_+_98881268 | 0.14 |
ENST00000254898.5 ENST00000524308.1 ENST00000522025.2 |
MATN2 |
matrilin 2 |
| chr7_+_128312346 | 0.14 |
ENST00000480462.1 ENST00000378704.3 ENST00000477515.1 |
FAM71F2 |
family with sequence similarity 71, member F2 |
| chr4_-_111119804 | 0.14 |
ENST00000394607.3 ENST00000302274.3 |
ELOVL6 |
ELOVL fatty acid elongase 6 |
| chrX_+_67913471 | 0.14 |
ENST00000374597.3 |
STARD8 |
StAR-related lipid transfer (START) domain containing 8 |
| chr19_+_45394477 | 0.14 |
ENST00000252487.5 ENST00000405636.2 ENST00000592434.1 ENST00000426677.2 ENST00000589649.1 |
TOMM40 |
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr19_-_40228657 | 0.14 |
ENST00000221804.4 |
CLC |
Charcot-Leyden crystal galectin |
| chr7_-_121944491 | 0.14 |
ENST00000331178.4 ENST00000427185.2 ENST00000442488.2 |
FEZF1 |
FEZ family zinc finger 1 |
| chr4_+_30723003 | 0.14 |
ENST00000543491.1 |
PCDH7 |
protocadherin 7 |
| chrX_-_70326455 | 0.13 |
ENST00000374251.5 |
CXorf65 |
chromosome X open reading frame 65 |
| chr2_-_192016316 | 0.13 |
ENST00000358470.4 ENST00000432798.1 ENST00000450994.1 |
STAT4 |
signal transducer and activator of transcription 4 |
| chr17_-_73285293 | 0.13 |
ENST00000582778.1 ENST00000581988.1 ENST00000579207.1 ENST00000583332.1 ENST00000416858.2 ENST00000442286.2 ENST00000580151.1 ENST00000580994.1 ENST00000584438.1 ENST00000320362.3 ENST00000580273.1 |
SLC25A19 |
solute carrier family 25 (mitochondrial thiamine pyrophosphate carrier), member 19 |
| chr2_+_85661918 | 0.13 |
ENST00000340326.2 |
SH2D6 |
SH2 domain containing 6 |
| chr1_-_11036272 | 0.13 |
ENST00000520253.1 |
C1orf127 |
chromosome 1 open reading frame 127 |
| chr17_+_44928946 | 0.13 |
ENST00000290015.2 ENST00000393461.2 |
WNT9B |
wingless-type MMTV integration site family, member 9B |
| chr22_+_30821732 | 0.13 |
ENST00000355143.4 |
MTFP1 |
mitochondrial fission process 1 |
| chr20_-_13971255 | 0.13 |
ENST00000284951.5 ENST00000378072.5 |
SEL1L2 |
sel-1 suppressor of lin-12-like 2 (C. elegans) |
| chr12_+_20963647 | 0.13 |
ENST00000381545.3 |
SLCO1B3 |
solute carrier organic anion transporter family, member 1B3 |
| chr12_+_20963632 | 0.13 |
ENST00000540853.1 ENST00000261196.2 |
SLCO1B3 |
solute carrier organic anion transporter family, member 1B3 |
| chr11_-_5531215 | 0.13 |
ENST00000311659.4 |
UBQLN3 |
ubiquilin 3 |
| chr12_+_6494285 | 0.12 |
ENST00000541102.1 |
LTBR |
lymphotoxin beta receptor (TNFR superfamily, member 3) |
| chr13_-_95131923 | 0.12 |
ENST00000377028.5 ENST00000446125.1 |
DCT |
dopachrome tautomerase |
| chr1_+_93544791 | 0.12 |
ENST00000545708.1 ENST00000540243.1 ENST00000370298.4 |
MTF2 |
metal response element binding transcription factor 2 |
| chr15_-_41408339 | 0.12 |
ENST00000401393.3 |
INO80 |
INO80 complex subunit |
| chr19_+_49838653 | 0.12 |
ENST00000598095.1 ENST00000426897.2 ENST00000323906.4 ENST00000535669.2 ENST00000597602.1 ENST00000595660.1 |
CD37 |
CD37 molecule |
| chr10_+_99332529 | 0.12 |
ENST00000455090.1 |
ANKRD2 |
ankyrin repeat domain 2 (stretch responsive muscle) |
| chr19_+_32836499 | 0.12 |
ENST00000311921.4 ENST00000544431.1 ENST00000355898.5 |
ZNF507 |
zinc finger protein 507 |
| chr1_-_85040090 | 0.12 |
ENST00000370630.5 |
CTBS |
chitobiase, di-N-acetyl- |
| chrX_+_114827818 | 0.12 |
ENST00000420625.2 |
PLS3 |
plastin 3 |
| chr22_+_23264766 | 0.11 |
ENST00000390331.2 |
IGLC7 |
immunoglobulin lambda constant 7 |
| chr17_-_41132410 | 0.11 |
ENST00000409446.3 ENST00000453594.1 ENST00000409399.1 ENST00000421990.2 |
PTGES3L PTGES3L-AARSD1 |
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr1_-_109940550 | 0.11 |
ENST00000256637.6 |
SORT1 |
sortilin 1 |
| chr12_-_114843889 | 0.11 |
ENST00000405440.2 |
TBX5 |
T-box 5 |
| chr16_-_74734672 | 0.11 |
ENST00000306247.7 ENST00000575686.1 |
MLKL |
mixed lineage kinase domain-like |
| chr7_+_112090483 | 0.11 |
ENST00000403825.3 ENST00000429071.1 |
IFRD1 |
interferon-related developmental regulator 1 |
| chr11_+_7534999 | 0.11 |
ENST00000528947.1 ENST00000299492.4 |
PPFIBP2 |
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr1_+_16010779 | 0.11 |
ENST00000375799.3 ENST00000375793.2 |
PLEKHM2 |
pleckstrin homology domain containing, family M (with RUN domain) member 2 |
| chr16_-_74734742 | 0.11 |
ENST00000308807.7 ENST00000573267.1 |
MLKL |
mixed lineage kinase domain-like |
| chr1_+_96457545 | 0.11 |
ENST00000413825.2 |
RP11-147C23.1 |
Uncharacterized protein |
| chr12_-_25102252 | 0.11 |
ENST00000261192.7 |
BCAT1 |
branched chain amino-acid transaminase 1, cytosolic |
| chr9_+_36572851 | 0.11 |
ENST00000298048.2 ENST00000538311.1 ENST00000536987.1 ENST00000545008.1 ENST00000536860.1 ENST00000536329.1 ENST00000541717.1 ENST00000543751.1 |
MELK |
maternal embryonic leucine zipper kinase |
| chr10_-_52645379 | 0.11 |
ENST00000395489.2 |
A1CF |
APOBEC1 complementation factor |
| chr3_-_48471454 | 0.11 |
ENST00000296440.6 ENST00000448774.2 |
PLXNB1 |
plexin B1 |
| chr18_-_53804580 | 0.10 |
ENST00000590484.1 ENST00000589293.1 ENST00000587904.1 ENST00000591974.1 |
RP11-456O19.4 |
RP11-456O19.4 |
| chr4_-_151936416 | 0.10 |
ENST00000510413.1 ENST00000507224.1 |
LRBA |
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr11_-_30607819 | 0.10 |
ENST00000448418.2 |
MPPED2 |
metallophosphoesterase domain containing 2 |
| chr12_+_56477093 | 0.10 |
ENST00000549672.1 ENST00000415288.2 |
ERBB3 |
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr22_+_17082732 | 0.10 |
ENST00000558085.2 ENST00000592918.1 ENST00000400593.2 ENST00000592107.1 ENST00000426585.1 ENST00000591299.1 |
TPTEP1 |
transmembrane phosphatase with tensin homology pseudogene 1 |
| chr2_+_32853093 | 0.10 |
ENST00000448773.1 ENST00000317907.4 |
TTC27 |
tetratricopeptide repeat domain 27 |
| chr9_+_84304628 | 0.10 |
ENST00000437181.1 |
RP11-154D17.1 |
RP11-154D17.1 |
| chr6_+_139456226 | 0.10 |
ENST00000367658.2 |
HECA |
headcase homolog (Drosophila) |
| chr1_+_241815577 | 0.10 |
ENST00000366552.2 ENST00000437684.2 |
WDR64 |
WD repeat domain 64 |
| chr3_-_79068594 | 0.10 |
ENST00000436010.2 |
ROBO1 |
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr12_+_95611569 | 0.10 |
ENST00000261219.6 ENST00000551472.1 ENST00000552821.1 |
VEZT |
vezatin, adherens junctions transmembrane protein |
| chr2_+_197577841 | 0.10 |
ENST00000409270.1 |
CCDC150 |
coiled-coil domain containing 150 |
| chr14_-_21492113 | 0.10 |
ENST00000554094.1 |
NDRG2 |
NDRG family member 2 |
| chr9_-_139305051 | 0.10 |
ENST00000371725.3 ENST00000298537.7 |
SDCCAG3 |
serologically defined colon cancer antigen 3 |
| chr3_-_127872625 | 0.10 |
ENST00000464873.1 |
RUVBL1 |
RuvB-like AAA ATPase 1 |
| chr10_+_80828774 | 0.10 |
ENST00000334512.5 |
ZMIZ1 |
zinc finger, MIZ-type containing 1 |
| chr12_+_32655110 | 0.10 |
ENST00000546442.1 ENST00000583694.1 |
FGD4 |
FYVE, RhoGEF and PH domain containing 4 |
| chr1_-_244006528 | 0.10 |
ENST00000336199.5 ENST00000263826.5 |
AKT3 |
v-akt murine thymoma viral oncogene homolog 3 |
| chr5_-_145483932 | 0.10 |
ENST00000311450.4 |
PLAC8L1 |
PLAC8-like 1 |
| chr1_+_45212051 | 0.10 |
ENST00000372222.3 |
KIF2C |
kinesin family member 2C |
| chr11_+_3876859 | 0.10 |
ENST00000300737.4 |
STIM1 |
stromal interaction molecule 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.6 | GO:0042335 | cuticle development(GO:0042335) |
| 0.3 | 1.1 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.2 | 0.7 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.2 | 0.6 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.2 | 0.6 | GO:0072428 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.2 | 0.6 | GO:0021823 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.2 | 0.5 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) cellular alkene metabolic process(GO:0043449) |
| 0.2 | 0.5 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.1 | 0.7 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.1 | 0.5 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 0.4 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.1 | 0.4 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.1 | 0.3 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 0.5 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.1 | 0.3 | GO:0097254 | renal tubular secretion(GO:0097254) |
| 0.1 | 0.5 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.1 | 0.4 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.1 | 0.2 | GO:2001302 | lipoxin biosynthetic process(GO:2001301) lipoxin A4 metabolic process(GO:2001302) lipoxin A4 biosynthetic process(GO:2001303) |
| 0.1 | 0.3 | GO:0070434 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 0.1 | 0.2 | GO:2000276 | negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.1 | 0.2 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.1 | 0.5 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 0.2 | GO:1900106 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.1 | 0.2 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.1 | 0.9 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.9 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.0 | 0.2 | GO:1902162 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.3 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 0.0 | 0.3 | GO:0032185 | septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.3 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.1 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.0 | 0.3 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.3 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.0 | 0.1 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.0 | 0.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.2 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.2 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.2 | GO:1903923 | protein processing in phagocytic vesicle(GO:1900756) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.3 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.6 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 2.1 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.2 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 0.2 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 | 0.6 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.2 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 | 0.2 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.2 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:0090156 | cellular sphingolipid homeostasis(GO:0090156) |
| 0.0 | 0.2 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.2 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.3 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.1 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.0 | 0.3 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.5 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.1 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.0 | 0.6 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.0 | 0.2 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.3 | GO:0015866 | ADP transport(GO:0015866) |
| 0.0 | 0.5 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.5 | GO:0044130 | negative regulation of growth of symbiont in host(GO:0044130) |
| 0.0 | 0.1 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.2 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.4 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.0 | 0.1 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.0 | 0.1 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.1 | GO:0019474 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.0 | 0.1 | GO:0009098 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.0 | 0.1 | GO:0035989 | tendon development(GO:0035989) |
| 0.0 | 0.7 | GO:0032757 | positive regulation of interleukin-8 production(GO:0032757) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:0042759 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.1 | GO:0002249 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.0 | 0.4 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.0 | GO:0007500 | mesodermal cell fate determination(GO:0007500) negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.0 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.1 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.2 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.2 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.2 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.0 | 0.6 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.1 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.0 | 0.1 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.2 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.1 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.3 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.1 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.1 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.1 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.0 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.1 | 0.3 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.1 | 0.2 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.1 | 0.2 | GO:0071751 | IgA immunoglobulin complex(GO:0071745) IgA immunoglobulin complex, circulating(GO:0071746) monomeric IgA immunoglobulin complex(GO:0071748) polymeric IgA immunoglobulin complex(GO:0071749) secretory IgA immunoglobulin complex(GO:0071751) |
| 0.1 | 0.2 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.2 | GO:0097513 | actomyosin contractile ring(GO:0005826) myosin II filament(GO:0097513) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.7 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.3 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.3 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.1 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.1 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.0 | 0.4 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.0 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.1 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.0 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.2 | 2.7 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.2 | 1.1 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.1 | 0.5 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.5 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.1 | 1.9 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.4 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.2 | GO:0047977 | linoleate 13S-lipoxygenase activity(GO:0016165) hepoxilin-epoxide hydrolase activity(GO:0047977) |
| 0.1 | 0.2 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.5 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.1 | 0.1 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.1 | 0.5 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.5 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.2 | GO:0004040 | amidase activity(GO:0004040) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.8 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.5 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.0 | 0.2 | GO:0060422 | peptidyl-dipeptidase inhibitor activity(GO:0060422) |
| 0.0 | 0.4 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) |
| 0.0 | 0.4 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.9 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.3 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.4 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.2 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.3 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.3 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.2 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.1 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.0 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.1 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.2 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.5 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.7 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.1 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.7 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.3 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.1 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.0 | 0.8 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.3 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.1 | GO:0039706 | co-receptor binding(GO:0039706) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 1.0 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.9 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.5 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.5 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.6 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.8 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.5 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.5 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.9 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.8 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.3 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.5 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.6 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.8 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 2.6 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.6 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.3 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.9 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |