Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
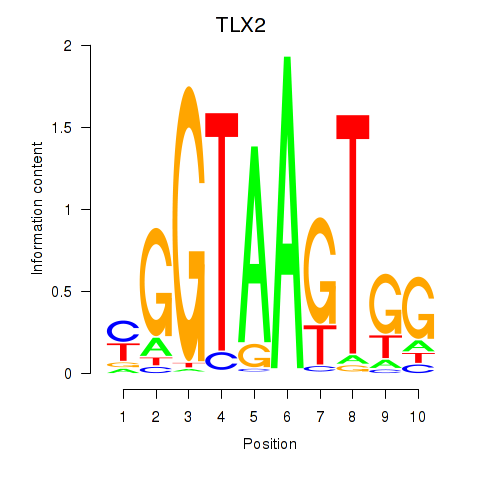
Results for TLX2
Z-value: 0.83
Transcription factors associated with TLX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TLX2
|
ENSG00000115297.9 | TLX2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TLX2 | hg19_v2_chr2_+_74741569_74741620 | -0.43 | 2.8e-01 | Click! |
Activity profile of TLX2 motif
Sorted Z-values of TLX2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TLX2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_86889769 | 1.21 |
ENST00000370565.4 |
CLCA2 |
chloride channel accessory 2 |
| chr11_+_69924639 | 1.17 |
ENST00000538023.1 ENST00000398543.2 |
ANO1 |
anoctamin 1, calcium activated chloride channel |
| chr15_+_41136586 | 1.07 |
ENST00000431806.1 |
SPINT1 |
serine peptidase inhibitor, Kunitz type 1 |
| chr1_-_120935894 | 1.05 |
ENST00000369383.4 ENST00000369384.4 |
FCGR1B |
Fc fragment of IgG, high affinity Ib, receptor (CD64) |
| chr8_+_86376081 | 1.00 |
ENST00000285379.5 |
CA2 |
carbonic anhydrase II |
| chr10_+_47746929 | 0.88 |
ENST00000340243.6 ENST00000374277.5 ENST00000449464.2 ENST00000538825.1 ENST00000335083.5 |
ANXA8L2 AL603965.1 |
annexin A8-like 2 Protein LOC100996760 |
| chr19_-_6767516 | 0.83 |
ENST00000245908.6 |
SH2D3A |
SH2 domain containing 3A |
| chr1_-_153363452 | 0.82 |
ENST00000368732.1 ENST00000368733.3 |
S100A8 |
S100 calcium binding protein A8 |
| chr6_-_138428613 | 0.76 |
ENST00000421351.3 |
PERP |
PERP, TP53 apoptosis effector |
| chr10_+_48255253 | 0.75 |
ENST00000357718.4 ENST00000344416.5 ENST00000456111.2 ENST00000374258.3 |
ANXA8 AL591684.1 |
annexin A8 Protein LOC100996760 |
| chr14_+_65171315 | 0.71 |
ENST00000394691.1 |
PLEKHG3 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr2_+_17721920 | 0.69 |
ENST00000295156.4 |
VSNL1 |
visinin-like 1 |
| chr16_-_68269971 | 0.69 |
ENST00000565858.1 |
ESRP2 |
epithelial splicing regulatory protein 2 |
| chr1_+_95286151 | 0.68 |
ENST00000467909.1 ENST00000422520.2 ENST00000532427.1 |
SLC44A3 |
solute carrier family 44, member 3 |
| chr5_+_131409476 | 0.63 |
ENST00000296871.2 |
CSF2 |
colony stimulating factor 2 (granulocyte-macrophage) |
| chr18_+_28956740 | 0.60 |
ENST00000308128.4 ENST00000359747.4 |
DSG4 |
desmoglein 4 |
| chr12_+_101188547 | 0.60 |
ENST00000546991.1 ENST00000392979.3 |
ANO4 |
anoctamin 4 |
| chr5_+_68710906 | 0.58 |
ENST00000325631.5 ENST00000454295.2 |
MARVELD2 |
MARVEL domain containing 2 |
| chr19_-_6767431 | 0.57 |
ENST00000437152.3 ENST00000597687.1 |
SH2D3A |
SH2 domain containing 3A |
| chr16_+_57406368 | 0.57 |
ENST00000006053.6 ENST00000563383.1 |
CX3CL1 |
chemokine (C-X3-C motif) ligand 1 |
| chr12_-_95611149 | 0.57 |
ENST00000549499.1 ENST00000343958.4 ENST00000546711.1 |
FGD6 |
FYVE, RhoGEF and PH domain containing 6 |
| chr1_+_153330322 | 0.57 |
ENST00000368738.3 |
S100A9 |
S100 calcium binding protein A9 |
| chr12_+_101188718 | 0.57 |
ENST00000299222.9 ENST00000392977.3 |
ANO4 |
anoctamin 4 |
| chr10_-_47173994 | 0.55 |
ENST00000414655.2 ENST00000545298.1 ENST00000359178.4 ENST00000358140.4 ENST00000503031.1 |
ANXA8L1 LINC00842 |
annexin A8-like 1 long intergenic non-protein coding RNA 842 |
| chr3_+_50192537 | 0.54 |
ENST00000002829.3 |
SEMA3F |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr3_+_50192499 | 0.52 |
ENST00000413852.1 |
SEMA3F |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr20_-_14318248 | 0.52 |
ENST00000378053.3 ENST00000341420.4 |
FLRT3 |
fibronectin leucine rich transmembrane protein 3 |
| chr12_+_56732658 | 0.52 |
ENST00000228534.4 |
IL23A |
interleukin 23, alpha subunit p19 |
| chr14_+_65171099 | 0.52 |
ENST00000247226.7 |
PLEKHG3 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr1_-_6526192 | 0.51 |
ENST00000377782.3 ENST00000351959.5 ENST00000356876.3 |
TNFRSF25 |
tumor necrosis factor receptor superfamily, member 25 |
| chr1_-_24469602 | 0.51 |
ENST00000270800.1 |
IL22RA1 |
interleukin 22 receptor, alpha 1 |
| chr1_+_152881014 | 0.50 |
ENST00000368764.3 ENST00000392667.2 |
IVL |
involucrin |
| chr1_+_62417957 | 0.50 |
ENST00000307297.7 ENST00000543708.1 |
INADL |
InaD-like (Drosophila) |
| chr3_+_124355944 | 0.50 |
ENST00000459915.1 |
KALRN |
kalirin, RhoGEF kinase |
| chr7_-_32111009 | 0.50 |
ENST00000396184.3 ENST00000396189.2 ENST00000321453.7 |
PDE1C |
phosphodiesterase 1C, calmodulin-dependent 70kDa |
| chr19_-_51512804 | 0.49 |
ENST00000594211.1 ENST00000376832.4 |
KLK9 |
kallikrein-related peptidase 9 |
| chr1_+_152486950 | 0.47 |
ENST00000368790.3 |
CRCT1 |
cysteine-rich C-terminal 1 |
| chr3_+_121796697 | 0.47 |
ENST00000482356.1 ENST00000393627.2 |
CD86 |
CD86 molecule |
| chr7_+_89783689 | 0.46 |
ENST00000297205.2 |
STEAP1 |
six transmembrane epithelial antigen of the prostate 1 |
| chr11_+_117947782 | 0.45 |
ENST00000522307.1 ENST00000523251.1 ENST00000437212.3 ENST00000522824.1 ENST00000522151.1 |
TMPRSS4 |
transmembrane protease, serine 4 |
| chr6_-_52109335 | 0.45 |
ENST00000336123.4 |
IL17F |
interleukin 17F |
| chr19_+_50084561 | 0.44 |
ENST00000246794.5 |
PRRG2 |
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr8_+_120220561 | 0.44 |
ENST00000276681.6 |
MAL2 |
mal, T-cell differentiation protein 2 (gene/pseudogene) |
| chr3_-_182880541 | 0.43 |
ENST00000470251.1 ENST00000265598.3 |
LAMP3 |
lysosomal-associated membrane protein 3 |
| chrX_-_45629661 | 0.42 |
ENST00000602507.1 ENST00000602461.1 |
RP6-99M1.2 |
RP6-99M1.2 |
| chr18_+_29077990 | 0.42 |
ENST00000261590.8 |
DSG2 |
desmoglein 2 |
| chr3_-_149051194 | 0.42 |
ENST00000470080.1 |
TM4SF18 |
transmembrane 4 L six family member 18 |
| chr12_-_71031220 | 0.41 |
ENST00000334414.6 |
PTPRB |
protein tyrosine phosphatase, receptor type, B |
| chrX_+_38420623 | 0.41 |
ENST00000378482.2 |
TSPAN7 |
tetraspanin 7 |
| chr12_-_71031185 | 0.40 |
ENST00000548122.1 ENST00000551525.1 ENST00000550358.1 |
PTPRB |
protein tyrosine phosphatase, receptor type, B |
| chr17_-_46507537 | 0.40 |
ENST00000336915.6 |
SKAP1 |
src kinase associated phosphoprotein 1 |
| chr16_+_82068830 | 0.40 |
ENST00000199936.4 |
HSD17B2 |
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr1_+_9711781 | 0.39 |
ENST00000536656.1 ENST00000377346.4 |
PIK3CD |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
| chr7_-_22539771 | 0.39 |
ENST00000406890.2 ENST00000424363.1 |
STEAP1B |
STEAP family member 1B |
| chr6_+_106546808 | 0.38 |
ENST00000369089.3 |
PRDM1 |
PR domain containing 1, with ZNF domain |
| chr14_+_61789382 | 0.38 |
ENST00000555082.1 |
PRKCH |
protein kinase C, eta |
| chr16_+_5121814 | 0.38 |
ENST00000262374.5 ENST00000586840.1 |
ALG1 |
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase |
| chr12_+_6494285 | 0.37 |
ENST00000541102.1 |
LTBR |
lymphotoxin beta receptor (TNFR superfamily, member 3) |
| chr11_+_117947724 | 0.37 |
ENST00000534111.1 |
TMPRSS4 |
transmembrane protease, serine 4 |
| chr1_-_6526167 | 0.37 |
ENST00000351748.3 ENST00000348333.3 |
TNFRSF25 |
tumor necrosis factor receptor superfamily, member 25 |
| chr15_-_34502197 | 0.36 |
ENST00000557877.1 |
KATNBL1 |
katanin p80 subunit B-like 1 |
| chr11_+_35160709 | 0.36 |
ENST00000415148.2 ENST00000433354.2 ENST00000449691.2 ENST00000437706.2 ENST00000360158.4 ENST00000428726.2 ENST00000526669.2 ENST00000433892.2 ENST00000278386.6 ENST00000434472.2 ENST00000352818.4 ENST00000442151.2 |
CD44 |
CD44 molecule (Indian blood group) |
| chr9_+_35673853 | 0.33 |
ENST00000378357.4 |
CA9 |
carbonic anhydrase IX |
| chr6_+_116692102 | 0.33 |
ENST00000359564.2 |
DSE |
dermatan sulfate epimerase |
| chr1_+_24646002 | 0.33 |
ENST00000356046.2 |
GRHL3 |
grainyhead-like 3 (Drosophila) |
| chr19_+_676385 | 0.33 |
ENST00000166139.4 |
FSTL3 |
follistatin-like 3 (secreted glycoprotein) |
| chr1_-_217262933 | 0.33 |
ENST00000359162.2 |
ESRRG |
estrogen-related receptor gamma |
| chr1_+_24645865 | 0.32 |
ENST00000342072.4 |
GRHL3 |
grainyhead-like 3 (Drosophila) |
| chr1_-_217262969 | 0.32 |
ENST00000361525.3 |
ESRRG |
estrogen-related receptor gamma |
| chr4_-_36246060 | 0.31 |
ENST00000303965.4 |
ARAP2 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr1_+_24645807 | 0.31 |
ENST00000361548.4 |
GRHL3 |
grainyhead-like 3 (Drosophila) |
| chr8_+_98900132 | 0.31 |
ENST00000520016.1 |
MATN2 |
matrilin 2 |
| chr17_-_76128488 | 0.30 |
ENST00000322914.3 |
TMC6 |
transmembrane channel-like 6 |
| chr4_+_41937131 | 0.30 |
ENST00000504986.1 ENST00000508448.1 ENST00000513702.1 ENST00000325094.5 |
TMEM33 |
transmembrane protein 33 |
| chr8_-_105601134 | 0.30 |
ENST00000276654.5 ENST00000424843.2 |
LRP12 |
low density lipoprotein receptor-related protein 12 |
| chr8_+_80523962 | 0.29 |
ENST00000518491.1 |
STMN2 |
stathmin-like 2 |
| chr10_-_103880209 | 0.29 |
ENST00000425280.1 |
LDB1 |
LIM domain binding 1 |
| chr21_-_16437255 | 0.29 |
ENST00000400199.1 ENST00000400202.1 |
NRIP1 |
nuclear receptor interacting protein 1 |
| chr3_+_118892362 | 0.29 |
ENST00000497685.1 ENST00000264234.3 |
UPK1B |
uroplakin 1B |
| chr1_+_160160346 | 0.28 |
ENST00000368078.3 |
CASQ1 |
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr21_-_16437126 | 0.28 |
ENST00000318948.4 |
NRIP1 |
nuclear receptor interacting protein 1 |
| chr12_+_9144626 | 0.28 |
ENST00000543895.1 |
KLRG1 |
killer cell lectin-like receptor subfamily G, member 1 |
| chr3_+_121774202 | 0.28 |
ENST00000469710.1 ENST00000493101.1 ENST00000330540.2 ENST00000264468.5 |
CD86 |
CD86 molecule |
| chr1_-_11042094 | 0.28 |
ENST00000377004.4 ENST00000377008.4 |
C1orf127 |
chromosome 1 open reading frame 127 |
| chr1_+_160160283 | 0.27 |
ENST00000368079.3 |
CASQ1 |
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr8_+_80523321 | 0.27 |
ENST00000518111.1 |
STMN2 |
stathmin-like 2 |
| chr6_+_84563295 | 0.27 |
ENST00000369687.1 |
RIPPLY2 |
ripply transcriptional repressor 2 |
| chr4_-_165305086 | 0.27 |
ENST00000507270.1 ENST00000514618.1 ENST00000503008.1 |
MARCH1 |
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr2_-_239197201 | 0.26 |
ENST00000254658.3 |
PER2 |
period circadian clock 2 |
| chr1_-_165324983 | 0.26 |
ENST00000367893.4 |
LMX1A |
LIM homeobox transcription factor 1, alpha |
| chr19_+_39421556 | 0.26 |
ENST00000407800.2 ENST00000402029.3 |
MRPS12 |
mitochondrial ribosomal protein S12 |
| chr13_-_50265570 | 0.26 |
ENST00000378270.5 ENST00000378284.2 ENST00000378272.5 ENST00000378268.1 ENST00000242827.6 ENST00000378282.5 |
EBPL |
emopamil binding protein-like |
| chr12_+_19358228 | 0.26 |
ENST00000424268.1 ENST00000543806.1 |
PLEKHA5 |
pleckstrin homology domain containing, family A member 5 |
| chr19_-_11688447 | 0.25 |
ENST00000590420.1 |
ACP5 |
acid phosphatase 5, tartrate resistant |
| chr12_-_120763739 | 0.25 |
ENST00000549767.1 |
PLA2G1B |
phospholipase A2, group IB (pancreas) |
| chr2_-_62733476 | 0.25 |
ENST00000335390.5 |
TMEM17 |
transmembrane protein 17 |
| chr2_-_72375167 | 0.25 |
ENST00000001146.2 |
CYP26B1 |
cytochrome P450, family 26, subfamily B, polypeptide 1 |
| chr20_-_50418947 | 0.25 |
ENST00000371539.3 |
SALL4 |
spalt-like transcription factor 4 |
| chr2_+_204801471 | 0.24 |
ENST00000316386.6 ENST00000435193.1 |
ICOS |
inducible T-cell co-stimulator |
| chr1_-_36947120 | 0.24 |
ENST00000361632.4 |
CSF3R |
colony stimulating factor 3 receptor (granulocyte) |
| chr10_-_50970322 | 0.23 |
ENST00000374103.4 |
OGDHL |
oxoglutarate dehydrogenase-like |
| chr14_-_67859422 | 0.23 |
ENST00000556532.1 |
PLEK2 |
pleckstrin 2 |
| chr1_-_25291475 | 0.23 |
ENST00000338888.3 ENST00000399916.1 |
RUNX3 |
runt-related transcription factor 3 |
| chr17_+_7211280 | 0.23 |
ENST00000419711.2 ENST00000571955.1 ENST00000573714.1 |
EIF5A |
eukaryotic translation initiation factor 5A |
| chr10_-_50970382 | 0.23 |
ENST00000419399.1 ENST00000432695.1 |
OGDHL |
oxoglutarate dehydrogenase-like |
| chr11_-_102323740 | 0.23 |
ENST00000398136.2 |
TMEM123 |
transmembrane protein 123 |
| chr11_+_67159416 | 0.23 |
ENST00000307980.2 ENST00000544620.1 |
RAD9A |
RAD9 homolog A (S. pombe) |
| chr19_-_36342739 | 0.22 |
ENST00000378910.5 ENST00000353632.6 |
NPHS1 |
nephrosis 1, congenital, Finnish type (nephrin) |
| chr16_-_28222797 | 0.22 |
ENST00000569951.1 ENST00000565698.1 |
XPO6 |
exportin 6 |
| chr1_+_36789335 | 0.22 |
ENST00000373137.2 |
RP11-268J15.5 |
RP11-268J15.5 |
| chr4_+_17812525 | 0.22 |
ENST00000251496.2 |
NCAPG |
non-SMC condensin I complex, subunit G |
| chr14_+_23340822 | 0.22 |
ENST00000359591.4 |
LRP10 |
low density lipoprotein receptor-related protein 10 |
| chr11_-_1036706 | 0.22 |
ENST00000421673.2 |
MUC6 |
mucin 6, oligomeric mucus/gel-forming |
| chr3_+_184098065 | 0.22 |
ENST00000348986.3 |
CHRD |
chordin |
| chrX_+_56259316 | 0.21 |
ENST00000468660.1 |
KLF8 |
Kruppel-like factor 8 |
| chr11_+_128563652 | 0.21 |
ENST00000527786.2 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr11_-_57089671 | 0.21 |
ENST00000532437.1 |
TNKS1BP1 |
tankyrase 1 binding protein 1, 182kDa |
| chr19_+_1067271 | 0.21 |
ENST00000536472.1 ENST00000590214.1 |
HMHA1 |
histocompatibility (minor) HA-1 |
| chr16_+_81478775 | 0.21 |
ENST00000537098.3 |
CMIP |
c-Maf inducing protein |
| chr8_+_133879193 | 0.21 |
ENST00000377869.1 ENST00000220616.4 |
TG |
thyroglobulin |
| chr16_-_88851618 | 0.21 |
ENST00000301015.9 |
PIEZO1 |
piezo-type mechanosensitive ion channel component 1 |
| chr22_+_31477296 | 0.21 |
ENST00000426927.1 ENST00000440425.1 ENST00000358743.1 ENST00000347557.2 ENST00000333137.7 |
SMTN |
smoothelin |
| chr8_-_145018905 | 0.21 |
ENST00000398774.2 |
PLEC |
plectin |
| chrX_-_31285042 | 0.21 |
ENST00000378680.2 ENST00000378723.3 |
DMD |
dystrophin |
| chr16_-_72206034 | 0.20 |
ENST00000537465.1 ENST00000237353.10 |
PMFBP1 |
polyamine modulated factor 1 binding protein 1 |
| chr4_-_75719896 | 0.20 |
ENST00000395743.3 |
BTC |
betacellulin |
| chr9_-_117692697 | 0.20 |
ENST00000223795.2 |
TNFSF8 |
tumor necrosis factor (ligand) superfamily, member 8 |
| chr10_-_50396425 | 0.20 |
ENST00000374148.1 |
C10orf128 |
chromosome 10 open reading frame 128 |
| chr4_-_68411275 | 0.20 |
ENST00000273853.6 |
CENPC |
centromere protein C |
| chr12_-_54785074 | 0.20 |
ENST00000338010.5 ENST00000550774.1 |
ZNF385A |
zinc finger protein 385A |
| chr10_-_44880491 | 0.20 |
ENST00000374426.2 ENST00000395795.4 ENST00000395794.2 ENST00000374429.2 ENST00000395793.3 ENST00000343575.6 |
CXCL12 |
chemokine (C-X-C motif) ligand 12 |
| chr9_-_128003606 | 0.20 |
ENST00000324460.6 |
HSPA5 |
heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) |
| chr15_-_34628951 | 0.20 |
ENST00000397707.2 ENST00000560611.1 |
SLC12A6 |
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr5_-_524445 | 0.20 |
ENST00000514375.1 ENST00000264938.3 |
SLC9A3 |
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 |
| chr1_+_205473720 | 0.20 |
ENST00000429964.2 ENST00000506784.1 ENST00000360066.2 |
CDK18 |
cyclin-dependent kinase 18 |
| chr12_-_322504 | 0.20 |
ENST00000424061.2 |
SLC6A12 |
solute carrier family 6 (neurotransmitter transporter), member 12 |
| chr1_+_231114795 | 0.20 |
ENST00000310256.2 ENST00000366658.2 ENST00000450711.1 ENST00000435927.1 |
ARV1 |
ARV1 homolog (S. cerevisiae) |
| chr20_-_47894936 | 0.19 |
ENST00000371754.4 |
ZNFX1 |
zinc finger, NFX1-type containing 1 |
| chr12_-_89918522 | 0.19 |
ENST00000529983.2 |
GALNT4 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4) |
| chr14_+_92980111 | 0.19 |
ENST00000216487.7 ENST00000557762.1 |
RIN3 |
Ras and Rab interactor 3 |
| chr13_+_113777105 | 0.19 |
ENST00000409306.1 ENST00000375551.3 ENST00000375559.3 |
F10 |
coagulation factor X |
| chr4_-_147443043 | 0.19 |
ENST00000394059.4 ENST00000502607.1 ENST00000335472.7 ENST00000432059.2 ENST00000394062.3 |
SLC10A7 |
solute carrier family 10, member 7 |
| chr1_-_31902614 | 0.19 |
ENST00000596131.1 |
AC114494.1 |
HCG1787699; Uncharacterized protein |
| chr21_+_43639211 | 0.18 |
ENST00000450121.1 ENST00000398449.3 ENST00000361802.2 |
ABCG1 |
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr11_-_93474645 | 0.18 |
ENST00000532455.1 |
TAF1D |
TATA box binding protein (TBP)-associated factor, RNA polymerase I, D, 41kDa |
| chr1_+_207494853 | 0.18 |
ENST00000367064.3 ENST00000367063.2 ENST00000391921.4 ENST00000367067.4 ENST00000314754.8 ENST00000367065.5 ENST00000391920.4 ENST00000367062.4 ENST00000343420.6 |
CD55 |
CD55 molecule, decay accelerating factor for complement (Cromer blood group) |
| chr2_+_191045562 | 0.18 |
ENST00000340623.4 |
C2orf88 |
chromosome 2 open reading frame 88 |
| chrX_-_72095622 | 0.18 |
ENST00000290273.5 |
DMRTC1 |
DMRT-like family C1 |
| chrX_-_73512411 | 0.18 |
ENST00000602576.1 ENST00000429124.1 |
FTX |
FTX transcript, XIST regulator (non-protein coding) |
| chr9_+_128024067 | 0.18 |
ENST00000461379.1 ENST00000394084.1 ENST00000394105.2 ENST00000470056.1 ENST00000394104.2 ENST00000265956.4 ENST00000394083.2 ENST00000495955.1 ENST00000467750.1 ENST00000297933.6 |
GAPVD1 |
GTPase activating protein and VPS9 domains 1 |
| chr2_+_219433588 | 0.18 |
ENST00000295701.5 |
RQCD1 |
RCD1 required for cell differentiation1 homolog (S. pombe) |
| chr20_-_1472029 | 0.18 |
ENST00000359801.3 |
SIRPB2 |
signal-regulatory protein beta 2 |
| chr7_+_39125365 | 0.18 |
ENST00000559001.1 ENST00000464276.2 |
POU6F2 |
POU class 6 homeobox 2 |
| chrX_-_72095808 | 0.18 |
ENST00000373529.5 |
DMRTC1 |
DMRT-like family C1 |
| chr3_-_42814708 | 0.17 |
ENST00000310232.6 |
CCDC13 |
coiled-coil domain containing 13 |
| chr17_-_46667594 | 0.17 |
ENST00000476342.1 ENST00000460160.1 ENST00000472863.1 |
HOXB3 |
homeobox B3 |
| chr6_-_31550192 | 0.17 |
ENST00000429299.2 ENST00000446745.2 |
LTB |
lymphotoxin beta (TNF superfamily, member 3) |
| chr3_-_107809816 | 0.17 |
ENST00000361309.5 ENST00000355354.7 |
CD47 |
CD47 molecule |
| chr10_+_73724123 | 0.17 |
ENST00000373115.4 |
CHST3 |
carbohydrate (chondroitin 6) sulfotransferase 3 |
| chr16_+_29674540 | 0.17 |
ENST00000436527.1 ENST00000360121.3 ENST00000449759.1 |
SPN QPRT |
sialophorin quinolinate phosphoribosyltransferase |
| chr18_+_55862622 | 0.17 |
ENST00000456173.2 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr12_+_104324112 | 0.16 |
ENST00000299767.5 |
HSP90B1 |
heat shock protein 90kDa beta (Grp94), member 1 |
| chr9_+_131799213 | 0.16 |
ENST00000358369.4 ENST00000406926.2 ENST00000277475.5 ENST00000450073.1 |
FAM73B |
family with sequence similarity 73, member B |
| chr7_-_33080506 | 0.16 |
ENST00000381626.2 ENST00000409467.1 ENST00000449201.1 |
NT5C3A |
5'-nucleotidase, cytosolic IIIA |
| chr17_+_61086917 | 0.16 |
ENST00000424789.2 ENST00000389520.4 |
TANC2 |
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chrX_+_24483338 | 0.16 |
ENST00000379162.4 ENST00000441463.2 |
PDK3 |
pyruvate dehydrogenase kinase, isozyme 3 |
| chr19_-_52035044 | 0.16 |
ENST00000359982.4 ENST00000436458.1 ENST00000425629.3 ENST00000391797.3 ENST00000343300.4 |
SIGLEC6 |
sialic acid binding Ig-like lectin 6 |
| chr2_+_160568978 | 0.16 |
ENST00000409175.1 ENST00000539065.1 ENST00000259050.4 ENST00000421037.1 |
MARCH7 |
membrane-associated ring finger (C3HC4) 7, E3 ubiquitin protein ligase |
| chr9_+_139847347 | 0.16 |
ENST00000371632.3 |
LCN12 |
lipocalin 12 |
| chr2_-_190445499 | 0.16 |
ENST00000261024.2 |
SLC40A1 |
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr1_+_1551220 | 0.16 |
ENST00000378712.1 |
MIB2 |
mindbomb E3 ubiquitin protein ligase 2 |
| chr7_+_93535817 | 0.16 |
ENST00000248572.5 |
GNGT1 |
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr15_+_89631647 | 0.16 |
ENST00000569550.1 ENST00000565066.1 ENST00000565973.1 |
ABHD2 |
abhydrolase domain containing 2 |
| chr3_+_186435137 | 0.16 |
ENST00000447445.1 |
KNG1 |
kininogen 1 |
| chr10_+_76871229 | 0.16 |
ENST00000372690.3 |
SAMD8 |
sterile alpha motif domain containing 8 |
| chr17_-_46667628 | 0.15 |
ENST00000498678.1 |
HOXB3 |
homeobox B3 |
| chr11_+_3819049 | 0.15 |
ENST00000396986.2 ENST00000300730.6 ENST00000532535.1 ENST00000396993.4 ENST00000396991.2 ENST00000532523.1 ENST00000459679.1 ENST00000464261.1 ENST00000464906.2 ENST00000464441.1 |
PGAP2 |
post-GPI attachment to proteins 2 |
| chr1_-_22263790 | 0.15 |
ENST00000374695.3 |
HSPG2 |
heparan sulfate proteoglycan 2 |
| chr20_-_13765526 | 0.15 |
ENST00000202816.1 |
ESF1 |
ESF1, nucleolar pre-rRNA processing protein, homolog (S. cerevisiae) |
| chr10_+_76871353 | 0.15 |
ENST00000542569.1 |
SAMD8 |
sterile alpha motif domain containing 8 |
| chr15_-_82555000 | 0.15 |
ENST00000557844.1 ENST00000359445.3 ENST00000268206.7 |
EFTUD1 |
elongation factor Tu GTP binding domain containing 1 |
| chr17_+_74734052 | 0.15 |
ENST00000590514.1 |
MFSD11 |
major facilitator superfamily domain containing 11 |
| chr1_+_16174280 | 0.15 |
ENST00000375759.3 |
SPEN |
spen family transcriptional repressor |
| chr4_+_140222609 | 0.15 |
ENST00000296543.5 ENST00000398947.1 |
NAA15 |
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
| chr1_-_156571254 | 0.15 |
ENST00000438976.2 ENST00000334588.7 ENST00000368232.4 ENST00000415314.2 |
GPATCH4 |
G patch domain containing 4 |
| chr3_-_149051444 | 0.15 |
ENST00000296059.2 |
TM4SF18 |
transmembrane 4 L six family member 18 |
| chr1_-_224033596 | 0.15 |
ENST00000391878.2 ENST00000343537.7 |
TP53BP2 |
tumor protein p53 binding protein, 2 |
| chr21_+_34398153 | 0.15 |
ENST00000382357.3 ENST00000430860.1 ENST00000333337.3 |
OLIG2 |
oligodendrocyte lineage transcription factor 2 |
| chr20_+_44746939 | 0.15 |
ENST00000372276.3 |
CD40 |
CD40 molecule, TNF receptor superfamily member 5 |
| chr1_+_16083154 | 0.15 |
ENST00000375771.1 |
FBLIM1 |
filamin binding LIM protein 1 |
| chr2_+_113403434 | 0.15 |
ENST00000272542.3 |
SLC20A1 |
solute carrier family 20 (phosphate transporter), member 1 |
| chr9_-_34397800 | 0.15 |
ENST00000297623.2 |
C9orf24 |
chromosome 9 open reading frame 24 |
| chr20_+_42544782 | 0.15 |
ENST00000423191.2 ENST00000372999.1 |
TOX2 |
TOX high mobility group box family member 2 |
| chr11_+_20620946 | 0.14 |
ENST00000525748.1 |
SLC6A5 |
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr1_+_236958554 | 0.14 |
ENST00000366577.5 ENST00000418145.2 |
MTR |
5-methyltetrahydrofolate-homocysteine methyltransferase |
| chr10_+_76871454 | 0.14 |
ENST00000372687.4 |
SAMD8 |
sterile alpha motif domain containing 8 |
| chr8_-_68658578 | 0.14 |
ENST00000518549.1 ENST00000297770.4 ENST00000297769.4 |
CPA6 |
carboxypeptidase A6 |
| chr19_-_13947099 | 0.14 |
ENST00000587762.1 |
MIR24-2 |
microRNA 24-2 |
| chr14_+_23938891 | 0.14 |
ENST00000408901.3 ENST00000397154.3 ENST00000555128.1 |
NGDN |
neuroguidin, EIF4E binding protein |
| chr6_+_44310421 | 0.14 |
ENST00000288390.2 |
SPATS1 |
spermatogenesis associated, serine-rich 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.3 | 1.3 | GO:0032641 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.2 | 0.6 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.2 | 0.6 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.2 | 0.7 | GO:1900138 | negative regulation of phospholipase A2 activity(GO:1900138) |
| 0.2 | 0.5 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.2 | 1.4 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 1.2 | GO:0061591 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylserine scrambling(GO:0061589) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.1 | 0.6 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.1 | 0.4 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.1 | 0.6 | GO:0045918 | positive regulation of interleukin-23 production(GO:0032747) negative regulation of cytolysis(GO:0045918) |
| 0.1 | 0.9 | GO:0035290 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) |
| 0.1 | 0.8 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.1 | 0.4 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.1 | 0.5 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.1 | 0.4 | GO:0003164 | His-Purkinje system development(GO:0003164) |
| 0.1 | 0.2 | GO:2001037 | tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.1 | 0.9 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 0.1 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.1 | 0.4 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) |
| 0.1 | 0.8 | GO:0032119 | sequestering of zinc ion(GO:0032119) |
| 0.1 | 0.4 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 0.2 | GO:0021919 | BMP signaling pathway involved in spinal cord dorsal/ventral patterning(GO:0021919) |
| 0.1 | 0.5 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.1 | 0.2 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.1 | 0.4 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.1 | 0.1 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.1 | 0.2 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.1 | 1.1 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.1 | 0.3 | GO:0051941 | regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 0.1 | 0.2 | GO:0070839 | divalent metal ion export(GO:0070839) |
| 0.1 | 1.0 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 0.2 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.2 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.6 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.0 | 0.3 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.2 | GO:0070352 | positive regulation of white fat cell proliferation(GO:0070352) |
| 0.0 | 0.5 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.0 | 0.1 | GO:0001694 | histamine biosynthetic process(GO:0001694) |
| 0.0 | 0.5 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.4 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.2 | GO:0048867 | ganglion mother cell fate determination(GO:0007402) stem cell fate determination(GO:0048867) |
| 0.0 | 0.4 | GO:0034351 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.0 | 0.2 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.0 | 0.2 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.3 | GO:0021546 | rhombomere development(GO:0021546) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.2 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.0 | 0.2 | GO:1902162 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.7 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.3 | GO:0033590 | response to cobalamin(GO:0033590) |
| 0.0 | 0.6 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.2 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.2 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.4 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.2 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.0 | 0.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.1 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.0 | 0.1 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.0 | 0.3 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.1 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.7 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 | 0.2 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 0.0 | 0.1 | GO:2000584 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.0 | 0.2 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.0 | 0.1 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.1 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.1 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.2 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.2 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.2 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.0 | 0.0 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.3 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.3 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.1 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.3 | GO:1904948 | midbrain dopaminergic neuron differentiation(GO:1904948) |
| 0.0 | 0.1 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.1 | GO:0060127 | pulmonary myocardium development(GO:0003350) subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) |
| 0.0 | 0.1 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.0 | 0.3 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.1 | GO:0071503 | response to heparin(GO:0071503) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.1 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.1 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.2 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.1 | GO:0019427 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate metabolic process(GO:0019541) propionate biosynthetic process(GO:0019542) |
| 0.0 | 0.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.2 | GO:0002517 | T cell tolerance induction(GO:0002517) |
| 0.0 | 0.4 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.4 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.1 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.3 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.6 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.0 | 0.2 | GO:0002643 | regulation of tolerance induction(GO:0002643) |
| 0.0 | 0.1 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.0 | 0.0 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.0 | 0.1 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.0 | 0.1 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.0 | 0.2 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.2 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.2 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.2 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.2 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.0 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.2 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.1 | GO:1902715 | secretory granule localization(GO:0032252) positive regulation of natural killer cell differentiation(GO:0032825) positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.0 | 0.0 | GO:0090202 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 | 0.1 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.2 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.0 | 0.4 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 0.0 | GO:0010725 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 0.0 | 0.2 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.0 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.2 | GO:0050650 | chondroitin sulfate biosynthetic process(GO:0030206) chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 0.0 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.1 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0097232 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.1 | 0.6 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 0.2 | GO:0005668 | RNA polymerase transcription factor SL1 complex(GO:0005668) |
| 0.1 | 0.4 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 1.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.6 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.5 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.4 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.2 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.3 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.2 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.2 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.3 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 1.1 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.0 | 0.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.4 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 1.3 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 1.0 | GO:0034707 | chloride channel complex(GO:0034707) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.2 | 1.4 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 1.0 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.1 | 1.0 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 0.8 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 2.5 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 1.6 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 0.4 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.1 | 0.4 | GO:0034437 | glycoprotein transporter activity(GO:0034437) |
| 0.1 | 0.6 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.1 | 0.5 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.5 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 0.4 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.1 | 0.7 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 0.5 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 0.3 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.1 | 0.2 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.1 | 0.5 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.2 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.4 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.0 | 0.2 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.2 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.0 | 1.5 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.5 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.1 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.0 | 0.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.2 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.8 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.3 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.1 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.6 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.2 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.6 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.4 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.7 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0022821 | potassium ion antiporter activity(GO:0022821) |
| 0.0 | 0.2 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.2 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.2 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.2 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.0 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.1 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.8 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.3 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 2.2 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.1 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.4 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.3 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.1 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 1.3 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.1 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.3 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.0 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.0 | 0.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.0 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.2 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.7 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.6 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 1.1 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.7 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.6 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 1.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.8 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.9 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 1.2 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 1.0 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.2 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 1.1 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |