Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for UAUUGCU
Z-value: 0.61
miRNA associated with seed UAUUGCU
| Name | miRBASE accession |
|---|---|
|
hsa-miR-137
|
MIMAT0000429 |
Activity profile of UAUUGCU motif
Sorted Z-values of UAUUGCU motif
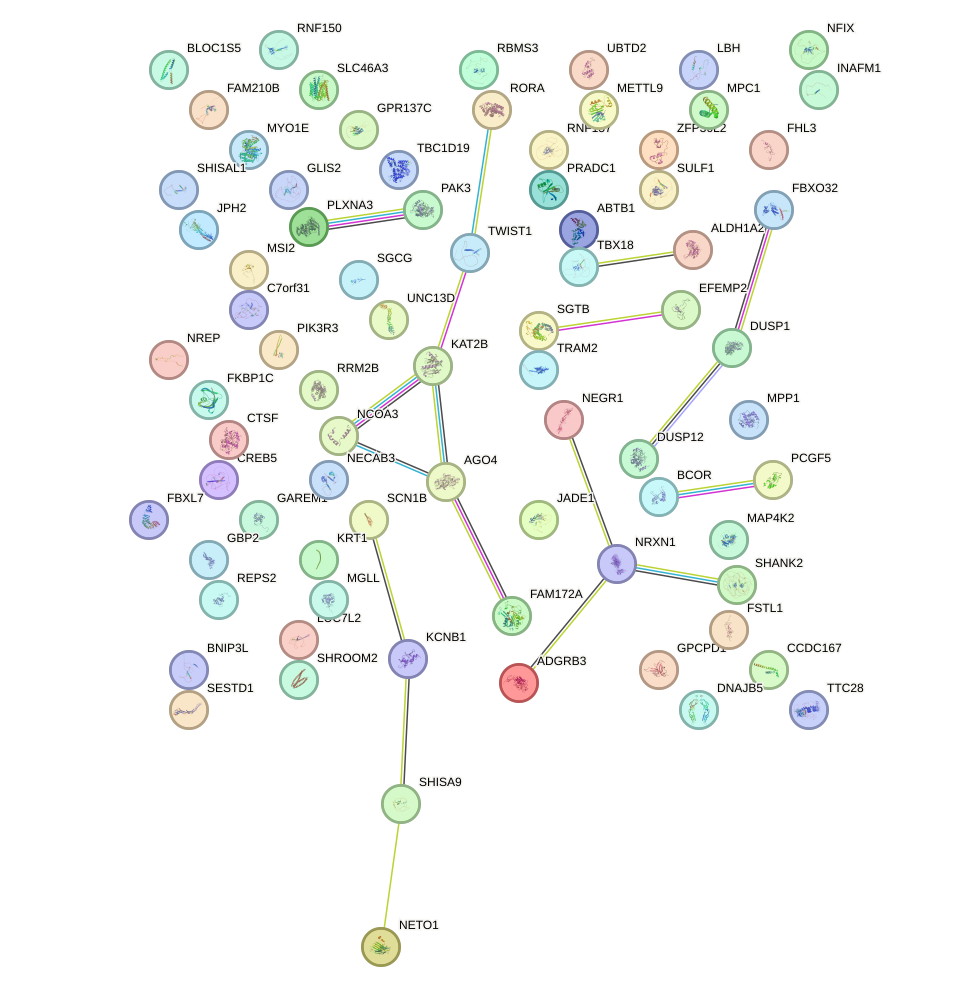
Network of associatons between targets according to the STRING database.
First level regulatory network of UAUUGCU
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_119799927 | 0.49 |
ENST00000407149.2 ENST00000379551.2 |
PRR16 |
proline rich 16 |
| chr1_+_213123915 | 0.35 |
ENST00000366968.4 ENST00000490792.1 |
VASH2 |
vasohibin 2 |
| chr5_-_111093406 | 0.32 |
ENST00000379671.3 |
NREP |
neuronal regeneration related protein |
| chrX_+_16964794 | 0.32 |
ENST00000357277.3 |
REPS2 |
RALBP1 associated Eps domain containing 2 |
| chr3_+_69812877 | 0.32 |
ENST00000457080.1 ENST00000328528.6 |
MITF |
microphthalmia-associated transcription factor |
| chr14_-_90085458 | 0.31 |
ENST00000345097.4 ENST00000555855.1 ENST00000555353.1 |
FOXN3 |
forkhead box N3 |
| chr5_-_172198190 | 0.31 |
ENST00000239223.3 |
DUSP1 |
dual specificity phosphatase 1 |
| chr1_-_72748417 | 0.31 |
ENST00000357731.5 |
NEGR1 |
neuronal growth regulator 1 |
| chr18_-_70535177 | 0.30 |
ENST00000327305.6 |
NETO1 |
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr13_+_32605437 | 0.30 |
ENST00000380250.3 |
FRY |
furry homolog (Drosophila) |
| chr20_-_48099182 | 0.29 |
ENST00000371741.4 |
KCNB1 |
potassium voltage-gated channel, Shab-related subfamily, member 1 |
| chr12_-_53074182 | 0.26 |
ENST00000252244.3 |
KRT1 |
keratin 1 |
| chr22_-_29075853 | 0.24 |
ENST00000397906.2 |
TTC28 |
tetratricopeptide repeat domain 28 |
| chr1_-_46598284 | 0.23 |
ENST00000423209.1 ENST00000262741.5 |
PIK3R3 |
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr1_-_232697304 | 0.22 |
ENST00000366630.1 |
SIPA1L2 |
signal-induced proliferation-associated 1 like 2 |
| chr19_+_10527449 | 0.19 |
ENST00000592685.1 ENST00000380702.2 |
PDE4A |
phosphodiesterase 4A, cAMP-specific |
| chr5_-_127873659 | 0.19 |
ENST00000262464.4 |
FBN2 |
fibrillin 2 |
| chrX_+_9754461 | 0.18 |
ENST00000380913.3 |
SHROOM2 |
shroom family member 2 |
| chr4_-_142053952 | 0.18 |
ENST00000515673.2 |
RNF150 |
ring finger protein 150 |
| chr20_-_42816206 | 0.18 |
ENST00000372980.3 |
JPH2 |
junctophilin 2 |
| chr3_+_20081515 | 0.17 |
ENST00000263754.4 |
KAT2B |
K(lysine) acetyltransferase 2B |
| chr11_-_70507901 | 0.17 |
ENST00000449833.2 ENST00000357171.3 ENST00000449116.2 |
SHANK2 |
SH3 and multiple ankyrin repeat domains 2 |
| chr5_+_133861790 | 0.17 |
ENST00000395003.1 |
PHF15 |
jade family PHD finger 2 |
| chr13_-_29292956 | 0.17 |
ENST00000266943.6 |
SLC46A3 |
solute carrier family 46, member 3 |
| chr20_+_54933971 | 0.17 |
ENST00000371384.3 ENST00000437418.1 |
FAM210B |
family with sequence similarity 210, member B |
| chr6_-_85474219 | 0.16 |
ENST00000369663.5 |
TBX18 |
T-box 18 |
| chr5_-_81046922 | 0.16 |
ENST00000514493.1 ENST00000320672.4 |
SSBP2 |
single-stranded DNA binding protein 2 |
| chr7_+_139026057 | 0.16 |
ENST00000541515.3 |
LUC7L2 |
LUC7-like 2 (S. cerevisiae) |
| chr5_-_93447333 | 0.16 |
ENST00000395965.3 ENST00000505869.1 ENST00000509163.1 |
FAM172A |
family with sequence similarity 172, member A |
| chr17_+_55333876 | 0.16 |
ENST00000284073.2 |
MSI2 |
musashi RNA-binding protein 2 |
| chr5_-_171711061 | 0.16 |
ENST00000393792.2 |
UBTD2 |
ubiquitin domain containing 2 |
| chr15_-_58358607 | 0.16 |
ENST00000249750.4 |
ALDH1A2 |
aldehyde dehydrogenase 1 family, member A2 |
| chr8_+_70378852 | 0.15 |
ENST00000525061.1 ENST00000458141.2 ENST00000260128.4 |
SULF1 |
sulfatase 1 |
| chr6_-_52441713 | 0.15 |
ENST00000182527.3 |
TRAM2 |
translocation associated membrane protein 2 |
| chr11_-_65640325 | 0.14 |
ENST00000307998.6 |
EFEMP2 |
EGF containing fibulin-like extracellular matrix protein 2 |
| chr11_-_64570706 | 0.14 |
ENST00000294066.2 ENST00000377350.3 |
MAP4K2 |
mitogen-activated protein kinase kinase kinase kinase 2 |
| chr1_-_38471156 | 0.14 |
ENST00000373016.3 |
FHL3 |
four and a half LIM domains 3 |
| chr12_-_31744031 | 0.14 |
ENST00000389082.5 |
DENND5B |
DENN/MADD domain containing 5B |
| chr16_+_12995468 | 0.14 |
ENST00000424107.3 ENST00000558583.1 ENST00000558318.1 |
SHISA9 |
shisa family member 9 |
| chr12_-_15942309 | 0.14 |
ENST00000544064.1 ENST00000543523.1 ENST00000536793.1 |
EPS8 |
epidermal growth factor receptor pathway substrate 8 |
| chr17_-_74236382 | 0.14 |
ENST00000592271.1 ENST00000319945.6 ENST00000269391.6 |
RNF157 |
ring finger protein 157 |
| chr3_-_120170052 | 0.14 |
ENST00000295633.3 |
FSTL1 |
follistatin-like 1 |
| chr16_+_4364762 | 0.14 |
ENST00000262366.3 |
GLIS2 |
GLIS family zinc finger 2 |
| chr3_+_29322803 | 0.14 |
ENST00000396583.3 ENST00000383767.2 |
RBMS3 |
RNA binding motif, single stranded interacting protein 3 |
| chr11_-_66336060 | 0.13 |
ENST00000310325.5 |
CTSF |
cathepsin F |
| chr1_-_89591749 | 0.13 |
ENST00000370466.3 |
GBP2 |
guanylate binding protein 2, interferon-inducible |
| chr7_-_19157248 | 0.13 |
ENST00000242261.5 |
TWIST1 |
twist family bHLH transcription factor 1 |
| chr19_+_35521572 | 0.12 |
ENST00000262631.5 |
SCN1B |
sodium channel, voltage-gated, type I, beta subunit |
| chr16_+_21610797 | 0.12 |
ENST00000358154.3 |
METTL9 |
methyltransferase like 9 |
| chr22_-_44708731 | 0.12 |
ENST00000381176.4 |
KIAA1644 |
KIAA1644 |
| chrX_-_154033793 | 0.12 |
ENST00000369534.3 ENST00000413259.3 |
MPP1 |
membrane protein, palmitoylated 1, 55kDa |
| chr19_+_47778119 | 0.12 |
ENST00000552360.2 |
PRR24 |
proline rich 24 |
| chr4_+_26585538 | 0.12 |
ENST00000264866.4 |
TBC1D19 |
TBC1 domain family, member 19 |
| chr1_-_47134101 | 0.12 |
ENST00000576409.1 |
ATPAF1 |
ATP synthase mitochondrial F1 complex assembly factor 1 |
| chr12_+_120105558 | 0.11 |
ENST00000229328.5 ENST00000541640.1 |
PRKAB1 |
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
| chr6_-_166796461 | 0.11 |
ENST00000360961.6 ENST00000341756.6 |
MPC1 |
mitochondrial pyruvate carrier 1 |
| chr20_-_32262165 | 0.11 |
ENST00000606690.1 ENST00000246190.6 ENST00000439478.1 ENST00000375238.4 |
NECAB3 |
N-terminal EF-hand calcium binding protein 3 |
| chr2_-_50574856 | 0.11 |
ENST00000342183.5 |
NRXN1 |
neurexin 1 |
| chr10_+_92980517 | 0.11 |
ENST00000336126.5 |
PCGF5 |
polycomb group ring finger 5 |
| chr18_-_30050395 | 0.11 |
ENST00000269209.6 ENST00000399218.4 |
GAREM |
GRB2 associated, regulator of MAPK1 |
| chr2_-_43453734 | 0.11 |
ENST00000282388.3 |
ZFP36L2 |
ZFP36 ring finger protein-like 2 |
| chrX_+_110339439 | 0.11 |
ENST00000372010.1 ENST00000519681.1 ENST00000372007.5 |
PAK3 |
p21 protein (Cdc42/Rac)-activated kinase 3 |
| chr8_-_124553437 | 0.11 |
ENST00000517956.1 ENST00000443022.2 |
FBXO32 |
F-box protein 32 |
| chr2_-_73460334 | 0.11 |
ENST00000258083.2 |
PRADC1 |
protease-associated domain containing 1 |
| chr7_+_28452130 | 0.11 |
ENST00000357727.2 |
CREB5 |
cAMP responsive element binding protein 5 |
| chr6_-_8064567 | 0.10 |
ENST00000543936.1 ENST00000397457.2 |
BLOC1S5 |
biogenesis of lysosomal organelles complex-1, subunit 5, muted |
| chr6_+_69345166 | 0.10 |
ENST00000370598.1 |
BAI3 |
brain-specific angiogenesis inhibitor 3 |
| chr4_+_129730779 | 0.10 |
ENST00000226319.6 |
PHF17 |
jade family PHD finger 1 |
| chr4_-_78740511 | 0.10 |
ENST00000504123.1 ENST00000264903.4 ENST00000515441.1 |
CNOT6L |
CCR4-NOT transcription complex, subunit 6-like |
| chr5_+_15500280 | 0.10 |
ENST00000504595.1 |
FBXL7 |
F-box and leucine-rich repeat protein 7 |
| chr5_-_65017921 | 0.10 |
ENST00000381007.4 |
SGTB |
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr3_-_127542021 | 0.10 |
ENST00000434178.2 |
MGLL |
monoglyceride lipase |
| chr11_+_2920951 | 0.10 |
ENST00000347936.2 |
SLC22A18 |
solute carrier family 22, member 18 |
| chr2_-_180129484 | 0.10 |
ENST00000428443.3 |
SESTD1 |
SEC14 and spectrin domains 1 |
| chr2_+_30454390 | 0.10 |
ENST00000395323.3 ENST00000406087.1 ENST00000404397.1 |
LBH |
limb bud and heart development |
| chr8_+_26240414 | 0.10 |
ENST00000380629.2 |
BNIP3L |
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr9_+_34990219 | 0.09 |
ENST00000541010.1 ENST00000454002.2 ENST00000545841.1 |
DNAJB5 |
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr20_-_1373726 | 0.09 |
ENST00000400137.4 |
FKBP1A |
FK506 binding protein 1A, 12kDa |
| chr20_-_5591626 | 0.09 |
ENST00000379019.4 |
GPCPD1 |
glycerophosphocholine phosphodiesterase GDE1 homolog (S. cerevisiae) |
| chr15_-_61521495 | 0.09 |
ENST00000335670.6 |
RORA |
RAR-related orphan receptor A |
| chr17_-_1395954 | 0.09 |
ENST00000359786.5 |
MYO1C |
myosin IC |
| chrX_+_153686614 | 0.09 |
ENST00000369682.3 |
PLXNA3 |
plexin A3 |
| chr6_+_63921351 | 0.09 |
ENST00000370659.1 |
FKBP1C |
FK506 binding protein 1C |
| chr14_+_53019822 | 0.09 |
ENST00000321662.6 |
GPR137C |
G protein-coupled receptor 137C |
| chr11_-_73309228 | 0.09 |
ENST00000356467.4 ENST00000064778.4 |
FAM168A |
family with sequence similarity 168, member A |
| chr7_-_25219897 | 0.09 |
ENST00000283905.3 ENST00000409280.1 ENST00000415598.1 |
C7orf31 |
chromosome 7 open reading frame 31 |
| chrX_-_39956656 | 0.09 |
ENST00000397354.3 ENST00000378444.4 |
BCOR |
BCL6 corepressor |
| chr8_-_103251274 | 0.09 |
ENST00000251810.3 |
RRM2B |
ribonucleotide reductase M2 B (TP53 inducible) |
| chr6_-_37467628 | 0.09 |
ENST00000373408.3 |
CCDC167 |
coiled-coil domain containing 167 |
| chr3_+_127391769 | 0.09 |
ENST00000393363.3 ENST00000232744.8 ENST00000453791.2 |
ABTB1 |
ankyrin repeat and BTB (POZ) domain containing 1 |
| chr1_+_36273743 | 0.09 |
ENST00000373210.3 |
AGO4 |
argonaute RISC catalytic component 4 |
| chr19_+_13135386 | 0.09 |
ENST00000360105.4 ENST00000588228.1 ENST00000591028.1 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr5_+_139944024 | 0.09 |
ENST00000323146.3 |
SLC35A4 |
solute carrier family 35, member A4 |
| chr13_+_23755054 | 0.09 |
ENST00000218867.3 |
SGCG |
sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein) |
| chr20_+_46130601 | 0.09 |
ENST00000341724.6 |
NCOA3 |
nuclear receptor coactivator 3 |
| chr18_+_11981427 | 0.09 |
ENST00000269159.3 |
IMPA2 |
inositol(myo)-1(or 4)-monophosphatase 2 |
| chr9_-_73736511 | 0.09 |
ENST00000377110.3 ENST00000377111.2 |
TRPM3 |
transient receptor potential cation channel, subfamily M, member 3 |
| chr17_-_19771216 | 0.09 |
ENST00000395544.4 |
ULK2 |
unc-51 like autophagy activating kinase 2 |
| chr6_-_111136513 | 0.09 |
ENST00000368911.3 |
CDK19 |
cyclin-dependent kinase 19 |
| chr3_+_88188254 | 0.08 |
ENST00000309495.5 |
ZNF654 |
zinc finger protein 654 |
| chr20_-_42939782 | 0.08 |
ENST00000396825.3 |
FITM2 |
fat storage-inducing transmembrane protein 2 |
| chr2_+_148778570 | 0.08 |
ENST00000407073.1 |
MBD5 |
methyl-CpG binding domain protein 5 |
| chrX_+_118108571 | 0.08 |
ENST00000304778.7 |
LONRF3 |
LON peptidase N-terminal domain and ring finger 3 |
| chr20_-_56284816 | 0.08 |
ENST00000395819.3 ENST00000341744.3 |
PMEPA1 |
prostate transmembrane protein, androgen induced 1 |
| chr9_+_112542572 | 0.08 |
ENST00000374530.3 |
PALM2-AKAP2 |
PALM2-AKAP2 readthrough |
| chr10_+_20105157 | 0.08 |
ENST00000377242.3 ENST00000377252.4 |
PLXDC2 |
plexin domain containing 2 |
| chr14_+_53196872 | 0.08 |
ENST00000442123.2 ENST00000354586.4 |
STYX |
serine/threonine/tyrosine interacting protein |
| chr12_+_65672423 | 0.08 |
ENST00000355192.3 ENST00000308259.5 ENST00000540804.1 ENST00000535664.1 ENST00000541189.1 |
MSRB3 |
methionine sulfoxide reductase B3 |
| chr5_-_133968529 | 0.08 |
ENST00000402673.2 |
SAR1B |
SAR1 homolog B (S. cerevisiae) |
| chr22_-_47134077 | 0.08 |
ENST00000541677.1 ENST00000216264.8 |
CERK |
ceramide kinase |
| chr15_-_55881135 | 0.08 |
ENST00000302000.6 |
PYGO1 |
pygopus family PHD finger 1 |
| chr2_-_200322723 | 0.08 |
ENST00000417098.1 |
SATB2 |
SATB homeobox 2 |
| chr5_-_107717058 | 0.08 |
ENST00000359660.5 |
FBXL17 |
F-box and leucine-rich repeat protein 17 |
| chr9_+_112810878 | 0.08 |
ENST00000434623.2 ENST00000374525.1 |
AKAP2 |
A kinase (PRKA) anchor protein 2 |
| chr5_+_110559784 | 0.08 |
ENST00000282356.4 |
CAMK4 |
calcium/calmodulin-dependent protein kinase IV |
| chr8_-_40755333 | 0.08 |
ENST00000297737.6 ENST00000315769.7 |
ZMAT4 |
zinc finger, matrin-type 4 |
| chr11_-_45687128 | 0.08 |
ENST00000308064.2 |
CHST1 |
carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 |
| chr10_+_104535994 | 0.08 |
ENST00000369889.4 |
WBP1L |
WW domain binding protein 1-like |
| chr17_-_1465924 | 0.08 |
ENST00000573231.1 ENST00000576722.1 ENST00000576761.1 ENST00000576010.2 ENST00000313486.7 ENST00000539476.1 |
PITPNA |
phosphatidylinositol transfer protein, alpha |
| chr1_+_61547894 | 0.08 |
ENST00000403491.3 |
NFIA |
nuclear factor I/A |
| chr21_-_44846999 | 0.08 |
ENST00000270162.6 |
SIK1 |
salt-inducible kinase 1 |
| chr2_+_120517174 | 0.08 |
ENST00000263708.2 |
PTPN4 |
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chrX_+_108780062 | 0.08 |
ENST00000372106.1 |
NXT2 |
nuclear transport factor 2-like export factor 2 |
| chr3_-_114790179 | 0.08 |
ENST00000462705.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr14_+_102829300 | 0.08 |
ENST00000359520.7 |
TECPR2 |
tectonin beta-propeller repeat containing 2 |
| chr11_-_124670550 | 0.07 |
ENST00000239614.4 |
MSANTD2 |
Myb/SANT-like DNA-binding domain containing 2 |
| chr21_+_45285050 | 0.07 |
ENST00000291572.8 |
AGPAT3 |
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr5_-_131132614 | 0.07 |
ENST00000307968.7 ENST00000307954.8 |
FNIP1 |
folliculin interacting protein 1 |
| chr14_-_35182994 | 0.07 |
ENST00000341223.3 |
CFL2 |
cofilin 2 (muscle) |
| chr20_+_30865429 | 0.07 |
ENST00000375712.3 |
KIF3B |
kinesin family member 3B |
| chr6_-_46293378 | 0.07 |
ENST00000330430.6 |
RCAN2 |
regulator of calcineurin 2 |
| chr5_+_118407053 | 0.07 |
ENST00000311085.8 ENST00000539542.1 |
DMXL1 |
Dmx-like 1 |
| chr1_-_1624083 | 0.07 |
ENST00000378662.1 ENST00000234800.6 |
SLC35E2B |
solute carrier family 35, member E2B |
| chr6_+_71998506 | 0.07 |
ENST00000370435.4 |
OGFRL1 |
opioid growth factor receptor-like 1 |
| chr10_-_46030841 | 0.07 |
ENST00000453424.2 |
MARCH8 |
membrane-associated ring finger (C3HC4) 8, E3 ubiquitin protein ligase |
| chr1_+_236305826 | 0.07 |
ENST00000366592.3 ENST00000366591.4 |
GPR137B |
G protein-coupled receptor 137B |
| chr11_-_44331679 | 0.07 |
ENST00000329255.3 |
ALX4 |
ALX homeobox 4 |
| chr16_+_66878814 | 0.07 |
ENST00000394069.3 |
CA7 |
carbonic anhydrase VII |
| chr7_+_138916231 | 0.07 |
ENST00000473989.3 ENST00000288561.8 |
UBN2 |
ubinuclein 2 |
| chr17_+_57642886 | 0.06 |
ENST00000251241.4 ENST00000451169.2 ENST00000425628.3 ENST00000584385.1 ENST00000580030.1 |
DHX40 |
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
| chr6_-_52926539 | 0.06 |
ENST00000350082.5 ENST00000356971.3 |
ICK |
intestinal cell (MAK-like) kinase |
| chr16_-_29910365 | 0.06 |
ENST00000346932.5 ENST00000350527.3 ENST00000537485.1 ENST00000568380.1 |
SEZ6L2 |
seizure related 6 homolog (mouse)-like 2 |
| chr1_+_112938803 | 0.06 |
ENST00000271277.6 |
CTTNBP2NL |
CTTNBP2 N-terminal like |
| chr12_-_54121212 | 0.06 |
ENST00000548263.1 ENST00000430117.2 ENST00000550804.1 ENST00000549173.1 ENST00000551900.1 ENST00000546619.1 ENST00000548177.1 ENST00000549349.1 |
CALCOCO1 |
calcium binding and coiled-coil domain 1 |
| chr8_-_139509065 | 0.06 |
ENST00000395297.1 |
FAM135B |
family with sequence similarity 135, member B |
| chr12_+_57482665 | 0.06 |
ENST00000300131.3 |
NAB2 |
NGFI-A binding protein 2 (EGR1 binding protein 2) |
| chr1_-_28969517 | 0.06 |
ENST00000263974.4 ENST00000373824.4 |
TAF12 |
TAF12 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 20kDa |
| chr3_+_37493610 | 0.06 |
ENST00000264741.5 |
ITGA9 |
integrin, alpha 9 |
| chr8_-_92053212 | 0.06 |
ENST00000285419.3 |
TMEM55A |
transmembrane protein 55A |
| chr16_+_1203194 | 0.06 |
ENST00000348261.5 ENST00000358590.4 |
CACNA1H |
calcium channel, voltage-dependent, T type, alpha 1H subunit |
| chr12_+_120972147 | 0.06 |
ENST00000325954.4 ENST00000542438.1 |
RNF10 |
ring finger protein 10 |
| chr9_-_125590818 | 0.06 |
ENST00000259467.4 |
PDCL |
phosducin-like |
| chr5_+_149887672 | 0.06 |
ENST00000261797.6 |
NDST1 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr6_-_48036363 | 0.06 |
ENST00000543600.1 ENST00000398738.2 ENST00000339488.4 |
PTCHD4 |
patched domain containing 4 |
| chr4_-_184580353 | 0.06 |
ENST00000326397.5 |
RWDD4 |
RWD domain containing 4 |
| chr16_+_11762270 | 0.06 |
ENST00000329565.5 |
SNN |
stannin |
| chr10_+_180987 | 0.06 |
ENST00000381591.1 |
ZMYND11 |
zinc finger, MYND-type containing 11 |
| chr20_+_32077880 | 0.06 |
ENST00000342704.6 ENST00000375279.2 |
CBFA2T2 |
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
| chr18_+_67956135 | 0.06 |
ENST00000397942.3 |
SOCS6 |
suppressor of cytokine signaling 6 |
| chr19_+_47104493 | 0.06 |
ENST00000291295.9 ENST00000597743.1 |
CALM3 |
calmodulin 3 (phosphorylase kinase, delta) |
| chr17_-_7832753 | 0.06 |
ENST00000303790.2 |
KCNAB3 |
potassium voltage-gated channel, shaker-related subfamily, beta member 3 |
| chr5_-_59189545 | 0.06 |
ENST00000340635.6 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
| chr5_-_38595498 | 0.06 |
ENST00000263409.4 |
LIFR |
leukemia inhibitory factor receptor alpha |
| chr19_+_16435625 | 0.06 |
ENST00000248071.5 ENST00000592003.1 |
KLF2 |
Kruppel-like factor 2 |
| chr16_-_20911641 | 0.06 |
ENST00000564349.1 ENST00000324344.4 |
ERI2 DCUN1D3 |
ERI1 exoribonuclease family member 2 DCN1, defective in cullin neddylation 1, domain containing 3 |
| chr22_+_50781723 | 0.06 |
ENST00000359139.3 ENST00000395741.3 ENST00000395744.3 |
PPP6R2 |
protein phosphatase 6, regulatory subunit 2 |
| chr1_+_43148059 | 0.06 |
ENST00000321358.7 ENST00000332220.6 |
YBX1 |
Y box binding protein 1 |
| chr11_+_24518723 | 0.05 |
ENST00000336930.6 ENST00000529015.1 ENST00000533227.1 |
LUZP2 |
leucine zipper protein 2 |
| chr1_-_222885770 | 0.05 |
ENST00000355727.2 ENST00000340020.6 |
AIDA |
axin interactor, dorsalization associated |
| chr1_+_154975110 | 0.05 |
ENST00000535420.1 ENST00000368426.3 |
ZBTB7B |
zinc finger and BTB domain containing 7B |
| chr1_+_36396677 | 0.05 |
ENST00000373191.4 ENST00000397828.2 |
AGO3 |
argonaute RISC catalytic component 3 |
| chr12_-_104532062 | 0.05 |
ENST00000240055.3 |
NFYB |
nuclear transcription factor Y, beta |
| chr14_+_74111578 | 0.05 |
ENST00000554113.1 ENST00000555631.2 ENST00000553645.2 ENST00000311089.3 ENST00000555919.3 ENST00000554339.1 ENST00000554871.1 |
DNAL1 |
dynein, axonemal, light chain 1 |
| chr2_-_197457335 | 0.05 |
ENST00000260983.3 |
HECW2 |
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr2_+_24714729 | 0.05 |
ENST00000406961.1 ENST00000405141.1 |
NCOA1 |
nuclear receptor coactivator 1 |
| chr4_-_66536057 | 0.05 |
ENST00000273854.3 |
EPHA5 |
EPH receptor A5 |
| chr4_-_17783135 | 0.05 |
ENST00000265018.3 |
FAM184B |
family with sequence similarity 184, member B |
| chr4_+_37892682 | 0.05 |
ENST00000508802.1 ENST00000261439.4 ENST00000402522.1 |
TBC1D1 |
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
| chr2_+_238875597 | 0.05 |
ENST00000272930.4 ENST00000448502.1 ENST00000416292.1 ENST00000409633.1 ENST00000414443.1 ENST00000409953.1 ENST00000409332.1 |
UBE2F |
ubiquitin-conjugating enzyme E2F (putative) |
| chr22_+_31090793 | 0.05 |
ENST00000332585.6 ENST00000382310.3 ENST00000446658.2 |
OSBP2 |
oxysterol binding protein 2 |
| chr20_+_39657454 | 0.05 |
ENST00000361337.2 |
TOP1 |
topoisomerase (DNA) I |
| chr8_-_53322303 | 0.05 |
ENST00000276480.7 |
ST18 |
suppression of tumorigenicity 18 (breast carcinoma) (zinc finger protein) |
| chr3_+_42695176 | 0.05 |
ENST00000232974.6 ENST00000457842.3 |
ZBTB47 |
zinc finger and BTB domain containing 47 |
| chr19_+_35759824 | 0.05 |
ENST00000343550.5 |
USF2 |
upstream transcription factor 2, c-fos interacting |
| chr11_+_12695944 | 0.05 |
ENST00000361905.4 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr7_-_21985489 | 0.05 |
ENST00000356195.5 ENST00000447180.1 ENST00000373934.4 ENST00000457951.1 |
CDCA7L |
cell division cycle associated 7-like |
| chr7_-_129592700 | 0.05 |
ENST00000472396.1 ENST00000355621.3 |
UBE2H |
ubiquitin-conjugating enzyme E2H |
| chr22_-_33454354 | 0.05 |
ENST00000358763.2 |
SYN3 |
synapsin III |
| chr12_+_104458235 | 0.05 |
ENST00000229330.4 |
HCFC2 |
host cell factor C2 |
| chr9_-_71155783 | 0.05 |
ENST00000377311.3 |
TMEM252 |
transmembrane protein 252 |
| chr9_+_137533615 | 0.05 |
ENST00000371817.3 |
COL5A1 |
collagen, type V, alpha 1 |
| chr2_+_136499179 | 0.05 |
ENST00000272638.9 |
UBXN4 |
UBX domain protein 4 |
| chr6_+_56819773 | 0.05 |
ENST00000370750.2 |
BEND6 |
BEN domain containing 6 |
| chr2_-_213403565 | 0.05 |
ENST00000342788.4 ENST00000436443.1 |
ERBB4 |
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 4 |
| chr12_+_69864129 | 0.05 |
ENST00000547219.1 ENST00000299293.2 ENST00000549921.1 ENST00000550316.1 ENST00000548154.1 ENST00000547414.1 ENST00000550389.1 ENST00000550937.1 ENST00000549092.1 ENST00000550169.1 |
FRS2 |
fibroblast growth factor receptor substrate 2 |
| chr16_-_31106048 | 0.05 |
ENST00000300851.6 |
VKORC1 |
vitamin K epoxide reductase complex, subunit 1 |
| chr9_-_36400213 | 0.05 |
ENST00000259605.6 ENST00000353739.4 |
RNF38 |
ring finger protein 38 |
| chr20_-_60982330 | 0.05 |
ENST00000279101.5 |
CABLES2 |
Cdk5 and Abl enzyme substrate 2 |
| chr12_-_39299406 | 0.05 |
ENST00000331366.5 |
CPNE8 |
copine VIII |
| chr2_-_101034070 | 0.05 |
ENST00000264249.3 |
CHST10 |
carbohydrate sulfotransferase 10 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.1 | 0.3 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 0.2 | GO:0060823 | canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060823) |
| 0.0 | 0.2 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.3 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.1 | GO:2000276 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.0 | 0.0 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.0 | 0.1 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.0 | 0.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.2 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.5 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.1 | GO:1902361 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 | 0.1 | GO:0060994 | regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) |
| 0.0 | 0.1 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.1 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.0 | 0.0 | GO:0035989 | tendon development(GO:0035989) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.2 | GO:0048864 | stem cell development(GO:0048864) |
| 0.0 | 0.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.2 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.0 | 0.2 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.0 | 0.1 | GO:1904674 | positive regulation of somatic stem cell population maintenance(GO:1904674) |
| 0.0 | 0.2 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.1 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.0 | 0.1 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.0 | 0.1 | GO:0046619 | optic placode formation(GO:0001743) optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.0 | 0.1 | GO:0075044 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.0 | 0.1 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.1 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.0 | 0.0 | GO:2000364 | regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 0.0 | 0.1 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 | 0.3 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.0 | GO:1901860 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) |
| 0.0 | 0.0 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.1 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.1 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.3 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 0.0 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.0 | 0.0 | GO:0071336 | submandibular salivary gland formation(GO:0060661) hair follicle cell proliferation(GO:0071335) regulation of hair follicle cell proliferation(GO:0071336) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.0 | 0.0 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 | 0.1 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.0 | 0.0 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.1 | GO:0035624 | receptor transactivation(GO:0035624) |
| 0.0 | 0.1 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.1 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.2 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.0 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.0 | 0.1 | GO:0010624 | regulation of Schwann cell proliferation(GO:0010624) negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.0 | 0.1 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 | 0.0 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.0 | GO:1903676 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.0 | 0.1 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.1 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 0.0 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.0 | GO:0060926 | cardiac pacemaker cell development(GO:0060926) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.0 | GO:2000705 | dense core granule biogenesis(GO:0061110) positive regulation of relaxation of cardiac muscle(GO:1901899) regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.1 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.0 | GO:1903422 | regulation of synaptic vesicle priming(GO:0010807) negative regulation of synaptic vesicle recycling(GO:1903422) regulation of spontaneous neurotransmitter secretion(GO:1904048) |
| 0.0 | 0.1 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0010265 | SCF complex assembly(GO:0010265) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.1 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.0 | 0.1 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.1 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.0 | 0.1 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.2 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.1 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.1 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.0 | GO:0016935 | glycine-gated chloride channel complex(GO:0016935) |
| 0.0 | 0.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.0 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.0 | 0.1 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.2 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0052832 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.0 | 0.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.0 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.0 | 0.2 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.0 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.0 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.0 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |