Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
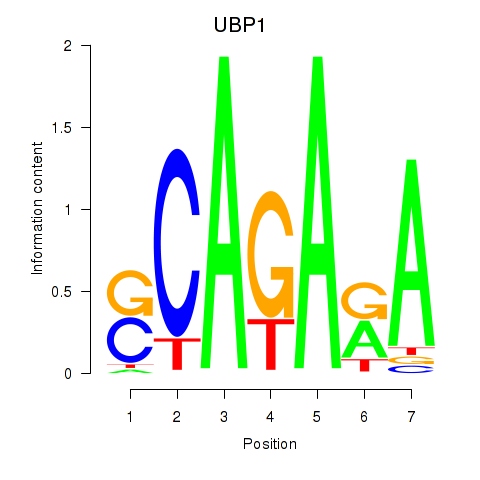
Results for UBP1
Z-value: 0.45
Transcription factors associated with UBP1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
UBP1
|
ENSG00000153560.7 | UBP1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| UBP1 | hg19_v2_chr3_-_33481835_33481905 | -0.44 | 2.8e-01 | Click! |
Activity profile of UBP1 motif
Sorted Z-values of UBP1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of UBP1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_92413353 | 0.73 |
ENST00000556154.1 |
FBLN5 |
fibulin 5 |
| chr14_-_92413727 | 0.62 |
ENST00000267620.10 |
FBLN5 |
fibulin 5 |
| chr5_-_111093759 | 0.57 |
ENST00000509979.1 ENST00000513100.1 ENST00000508161.1 ENST00000455559.2 |
NREP |
neuronal regeneration related protein |
| chr4_-_70626314 | 0.52 |
ENST00000510821.1 |
SULT1B1 |
sulfotransferase family, cytosolic, 1B, member 1 |
| chr3_-_100712352 | 0.52 |
ENST00000471714.1 ENST00000284322.5 |
ABI3BP |
ABI family, member 3 (NESH) binding protein |
| chr4_-_57524061 | 0.32 |
ENST00000508121.1 |
HOPX |
HOP homeobox |
| chr1_-_168698433 | 0.30 |
ENST00000367817.3 |
DPT |
dermatopontin |
| chr21_+_43823983 | 0.23 |
ENST00000291535.6 ENST00000450356.1 ENST00000319294.6 ENST00000398367.1 |
UBASH3A |
ubiquitin associated and SH3 domain containing A |
| chr5_-_41510656 | 0.23 |
ENST00000377801.3 |
PLCXD3 |
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr3_+_29323043 | 0.19 |
ENST00000452462.1 ENST00000456853.1 |
RBMS3 |
RNA binding motif, single stranded interacting protein 3 |
| chr16_+_29823427 | 0.19 |
ENST00000358758.7 ENST00000567659.1 ENST00000572820.1 |
PRRT2 |
proline-rich transmembrane protein 2 |
| chr8_+_77593448 | 0.19 |
ENST00000521891.2 |
ZFHX4 |
zinc finger homeobox 4 |
| chr9_-_35685452 | 0.18 |
ENST00000607559.1 |
TPM2 |
tropomyosin 2 (beta) |
| chr12_+_100041527 | 0.16 |
ENST00000324341.1 |
FAM71C |
family with sequence similarity 71, member C |
| chr11_-_134123142 | 0.15 |
ENST00000392595.2 ENST00000341541.3 ENST00000352327.5 ENST00000392594.3 |
THYN1 |
thymocyte nuclear protein 1 |
| chrX_-_65253506 | 0.15 |
ENST00000427538.1 |
VSIG4 |
V-set and immunoglobulin domain containing 4 |
| chr22_+_22730353 | 0.14 |
ENST00000390296.2 |
IGLV5-45 |
immunoglobulin lambda variable 5-45 |
| chr4_-_159080806 | 0.14 |
ENST00000590648.1 |
FAM198B |
family with sequence similarity 198, member B |
| chr16_-_67970990 | 0.13 |
ENST00000358514.4 |
PSMB10 |
proteasome (prosome, macropain) subunit, beta type, 10 |
| chr4_-_76861392 | 0.13 |
ENST00000505594.1 |
NAAA |
N-acylethanolamine acid amidase |
| chr19_-_58459039 | 0.12 |
ENST00000282308.3 ENST00000598928.1 |
ZNF256 |
zinc finger protein 256 |
| chr1_-_182369751 | 0.12 |
ENST00000367565.1 |
TEDDM1 |
transmembrane epididymal protein 1 |
| chr1_-_182641367 | 0.12 |
ENST00000508450.1 |
RGS8 |
regulator of G-protein signaling 8 |
| chr5_-_93447333 | 0.11 |
ENST00000395965.3 ENST00000505869.1 ENST00000509163.1 |
FAM172A |
family with sequence similarity 172, member A |
| chr22_+_38071615 | 0.11 |
ENST00000215909.5 |
LGALS1 |
lectin, galactoside-binding, soluble, 1 |
| chr7_+_142498725 | 0.11 |
ENST00000466254.1 |
TRBC2 |
T cell receptor beta constant 2 |
| chr22_+_22707260 | 0.10 |
ENST00000390293.1 |
IGLV5-48 |
immunoglobulin lambda variable 5-48 (non-functional) |
| chr11_+_7506713 | 0.09 |
ENST00000329293.3 ENST00000534244.1 |
OLFML1 |
olfactomedin-like 1 |
| chr11_+_10326612 | 0.09 |
ENST00000534464.1 ENST00000530439.1 ENST00000524948.1 ENST00000528655.1 ENST00000526492.1 ENST00000525063.1 |
ADM |
adrenomedullin |
| chr5_+_150051149 | 0.09 |
ENST00000523553.1 |
MYOZ3 |
myozenin 3 |
| chr3_-_56502375 | 0.09 |
ENST00000288221.6 |
ERC2 |
ELKS/RAB6-interacting/CAST family member 2 |
| chr12_-_123717643 | 0.09 |
ENST00000541437.1 ENST00000606320.1 |
MPHOSPH9 |
M-phase phosphoprotein 9 |
| chr13_-_23949671 | 0.08 |
ENST00000402364.1 |
SACS |
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chr11_+_124824000 | 0.08 |
ENST00000529051.1 ENST00000344762.5 |
CCDC15 |
coiled-coil domain containing 15 |
| chr11_-_60623437 | 0.08 |
ENST00000332539.4 |
PTGDR2 |
prostaglandin D2 receptor 2 |
| chrX_-_70474910 | 0.07 |
ENST00000373988.1 ENST00000373998.1 |
ZMYM3 |
zinc finger, MYM-type 3 |
| chrX_+_78426469 | 0.07 |
ENST00000276077.1 |
GPR174 |
G protein-coupled receptor 174 |
| chr2_+_170550944 | 0.07 |
ENST00000359744.3 ENST00000438838.1 ENST00000438710.1 ENST00000449906.1 ENST00000498202.2 ENST00000272797.4 |
PHOSPHO2 KLHL23 |
phosphatase, orphan 2 kelch-like family member 23 |
| chr1_-_153599732 | 0.07 |
ENST00000392623.1 |
S100A13 |
S100 calcium binding protein A13 |
| chr1_-_157014865 | 0.07 |
ENST00000361409.2 |
ARHGEF11 |
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr2_+_228678550 | 0.06 |
ENST00000409189.3 ENST00000358813.4 |
CCL20 |
chemokine (C-C motif) ligand 20 |
| chr12_+_9067123 | 0.06 |
ENST00000543824.1 |
PHC1 |
polyhomeotic homolog 1 (Drosophila) |
| chr1_-_202129704 | 0.06 |
ENST00000476061.1 ENST00000544762.1 ENST00000467283.1 ENST00000464870.1 ENST00000435759.2 ENST00000486116.1 ENST00000543735.1 ENST00000308986.5 ENST00000477625.1 |
PTPN7 |
protein tyrosine phosphatase, non-receptor type 7 |
| chr1_-_114302086 | 0.06 |
ENST00000369604.1 ENST00000357783.2 |
PHTF1 |
putative homeodomain transcription factor 1 |
| chr1_+_48688357 | 0.06 |
ENST00000533824.1 ENST00000438567.2 ENST00000236495.5 ENST00000420136.2 |
SLC5A9 |
solute carrier family 5 (sodium/sugar cotransporter), member 9 |
| chr19_-_59066327 | 0.05 |
ENST00000596708.1 ENST00000601220.1 ENST00000597848.1 |
CHMP2A |
charged multivesicular body protein 2A |
| chr1_-_150208320 | 0.05 |
ENST00000534220.1 |
ANP32E |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr6_+_29429217 | 0.05 |
ENST00000396792.2 |
OR2H1 |
olfactory receptor, family 2, subfamily H, member 1 |
| chr1_+_41174988 | 0.05 |
ENST00000372652.1 |
NFYC |
nuclear transcription factor Y, gamma |
| chr1_-_202129105 | 0.05 |
ENST00000367279.4 |
PTPN7 |
protein tyrosine phosphatase, non-receptor type 7 |
| chr12_+_123717967 | 0.05 |
ENST00000536130.1 ENST00000546132.1 |
C12orf65 |
chromosome 12 open reading frame 65 |
| chr2_+_162087577 | 0.05 |
ENST00000439442.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr5_+_95998070 | 0.04 |
ENST00000421689.2 ENST00000510756.1 ENST00000512620.1 |
CAST |
calpastatin |
| chr10_-_27443155 | 0.04 |
ENST00000427324.1 ENST00000326799.3 |
YME1L1 |
YME1-like 1 ATPase |
| chr8_-_66474884 | 0.04 |
ENST00000520902.1 |
CTD-3025N20.2 |
CTD-3025N20.2 |
| chr1_-_150208498 | 0.04 |
ENST00000314136.8 |
ANP32E |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr6_+_6588316 | 0.04 |
ENST00000379953.2 |
LY86 |
lymphocyte antigen 86 |
| chr1_-_150208412 | 0.04 |
ENST00000532744.1 ENST00000369114.5 ENST00000369115.2 ENST00000369116.4 |
ANP32E |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr1_-_152332480 | 0.04 |
ENST00000388718.5 |
FLG2 |
filaggrin family member 2 |
| chr1_-_150208291 | 0.04 |
ENST00000533654.1 |
ANP32E |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr1_-_150208363 | 0.04 |
ENST00000436748.2 |
ANP32E |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chrX_+_74493896 | 0.03 |
ENST00000373383.4 ENST00000373379.1 |
UPRT |
uracil phosphoribosyltransferase (FUR1) homolog (S. cerevisiae) |
| chr10_-_27443294 | 0.03 |
ENST00000396296.3 ENST00000375972.3 ENST00000376016.3 ENST00000491542.2 |
YME1L1 |
YME1-like 1 ATPase |
| chr9_-_6015607 | 0.03 |
ENST00000259569.5 |
RANBP6 |
RAN binding protein 6 |
| chr5_+_122847781 | 0.03 |
ENST00000395412.1 ENST00000395411.1 ENST00000345990.4 |
CSNK1G3 |
casein kinase 1, gamma 3 |
| chr14_+_75988768 | 0.03 |
ENST00000286639.6 |
BATF |
basic leucine zipper transcription factor, ATF-like |
| chr15_-_77712477 | 0.03 |
ENST00000560626.2 |
PEAK1 |
pseudopodium-enriched atypical kinase 1 |
| chr15_+_34638066 | 0.03 |
ENST00000333756.4 |
NUTM1 |
NUT midline carcinoma, family member 1 |
| chr6_+_31555045 | 0.03 |
ENST00000396101.3 ENST00000490742.1 |
LST1 |
leukocyte specific transcript 1 |
| chr12_+_123717458 | 0.03 |
ENST00000253233.1 |
C12orf65 |
chromosome 12 open reading frame 65 |
| chr1_+_63989004 | 0.03 |
ENST00000371088.4 |
EFCAB7 |
EF-hand calcium binding domain 7 |
| chr11_-_11374904 | 0.03 |
ENST00000528848.2 |
CSNK2A3 |
casein kinase 2, alpha 3 polypeptide |
| chr19_-_51538148 | 0.03 |
ENST00000319590.4 ENST00000250351.4 |
KLK12 |
kallikrein-related peptidase 12 |
| chr4_-_100815525 | 0.02 |
ENST00000226522.8 ENST00000499666.2 |
LAMTOR3 |
late endosomal/lysosomal adaptor, MAPK and MTOR activator 3 |
| chr7_+_142829162 | 0.02 |
ENST00000291009.3 |
PIP |
prolactin-induced protein |
| chr4_+_156588115 | 0.02 |
ENST00000455639.2 |
GUCY1A3 |
guanylate cyclase 1, soluble, alpha 3 |
| chr1_-_185126037 | 0.02 |
ENST00000367506.5 ENST00000367504.3 |
TRMT1L |
tRNA methyltransferase 1 homolog (S. cerevisiae)-like |
| chr5_-_131329918 | 0.02 |
ENST00000357096.1 ENST00000431707.1 ENST00000434099.1 |
ACSL6 |
acyl-CoA synthetase long-chain family member 6 |
| chr1_-_144995074 | 0.02 |
ENST00000534536.1 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chr22_+_22385332 | 0.02 |
ENST00000390282.2 |
IGLV4-69 |
immunoglobulin lambda variable 4-69 |
| chr2_-_2334888 | 0.02 |
ENST00000428368.2 ENST00000399161.2 |
MYT1L |
myelin transcription factor 1-like |
| chr5_-_132200477 | 0.02 |
ENST00000296875.2 |
GDF9 |
growth differentiation factor 9 |
| chr20_+_59654146 | 0.01 |
ENST00000441660.1 |
RP5-827L5.1 |
RP5-827L5.1 |
| chr15_+_81475047 | 0.01 |
ENST00000559388.1 |
IL16 |
interleukin 16 |
| chr1_+_206223941 | 0.01 |
ENST00000367126.4 |
AVPR1B |
arginine vasopressin receptor 1B |
| chr7_+_132937820 | 0.01 |
ENST00000393161.2 ENST00000253861.4 |
EXOC4 |
exocyst complex component 4 |
| chr4_-_168155730 | 0.01 |
ENST00000502330.1 ENST00000357154.3 |
SPOCK3 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr6_-_169364429 | 0.01 |
ENST00000444586.1 |
RP3-495K2.3 |
RP3-495K2.3 |
| chr1_-_150669604 | 0.01 |
ENST00000427665.1 ENST00000540514.1 |
GOLPH3L |
golgi phosphoprotein 3-like |
| chr17_-_48450534 | 0.01 |
ENST00000503633.1 ENST00000442592.3 ENST00000225969.4 |
MRPL27 |
mitochondrial ribosomal protein L27 |
| chr17_-_5138099 | 0.01 |
ENST00000571800.1 ENST00000574081.1 ENST00000399600.4 ENST00000574297.1 |
SCIMP |
SLP adaptor and CSK interacting membrane protein |
| chr11_+_128761220 | 0.01 |
ENST00000529694.1 |
KCNJ5 |
potassium inwardly-rectifying channel, subfamily J, member 5 |
| chr19_-_8070474 | 0.00 |
ENST00000407627.2 ENST00000593807.1 |
ELAVL1 |
ELAV like RNA binding protein 1 |
| chr4_-_47839966 | 0.00 |
ENST00000273857.4 ENST00000505909.1 ENST00000502252.1 |
CORIN |
corin, serine peptidase |
| chr1_-_144995002 | 0.00 |
ENST00000369356.4 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.1 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.0 | 0.5 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.1 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.0 | 0.1 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.0 | 0.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.3 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.1 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.2 | GO:0000812 | Swr1 complex(GO:0000812) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0030395 | lactose binding(GO:0030395) |
| 0.0 | 0.1 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.0 | 0.5 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |