Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
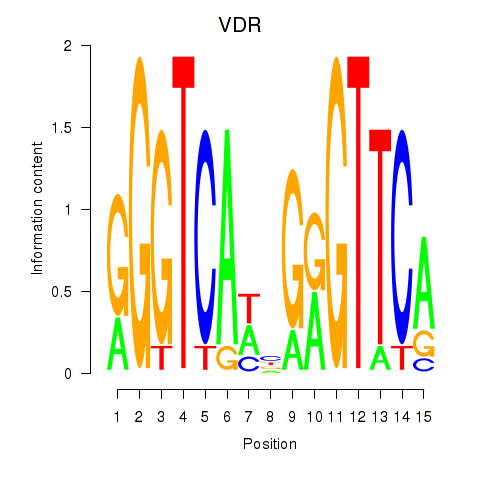
Results for VDR
Z-value: 0.59
Transcription factors associated with VDR
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
VDR
|
ENSG00000111424.6 | VDR |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| VDR | hg19_v2_chr12_-_48298785_48298828 | -0.38 | 3.5e-01 | Click! |
Activity profile of VDR motif
Sorted Z-values of VDR motif
Network of associatons between targets according to the STRING database.
First level regulatory network of VDR
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_133050726 | 0.68 |
ENST00000595994.1 |
MUC8 |
mucin 8 |
| chr3_+_157154578 | 0.55 |
ENST00000295927.3 |
PTX3 |
pentraxin 3, long |
| chr7_-_45960850 | 0.52 |
ENST00000381083.4 ENST00000381086.5 ENST00000275521.6 |
IGFBP3 |
insulin-like growth factor binding protein 3 |
| chr13_-_38172863 | 0.48 |
ENST00000541481.1 ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN |
periostin, osteoblast specific factor |
| chr3_+_45071622 | 0.42 |
ENST00000428034.1 |
CLEC3B |
C-type lectin domain family 3, member B |
| chr3_+_54157480 | 0.38 |
ENST00000490478.1 |
CACNA2D3 |
calcium channel, voltage-dependent, alpha 2/delta subunit 3 |
| chr8_-_13134045 | 0.27 |
ENST00000512044.2 |
DLC1 |
deleted in liver cancer 1 |
| chr13_-_33859819 | 0.27 |
ENST00000336934.5 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
| chr13_+_113656022 | 0.26 |
ENST00000423482.2 |
MCF2L |
MCF.2 cell line derived transforming sequence-like |
| chr19_-_54804173 | 0.26 |
ENST00000391744.3 ENST00000251390.3 |
LILRA3 |
leukocyte immunoglobulin-like receptor, subfamily A (without TM domain), member 3 |
| chr8_+_17434689 | 0.24 |
ENST00000398074.3 |
PDGFRL |
platelet-derived growth factor receptor-like |
| chr1_-_241799232 | 0.23 |
ENST00000366553.1 |
CHML |
choroideremia-like (Rab escort protein 2) |
| chr21_+_25801041 | 0.21 |
ENST00000453784.2 ENST00000423581.1 |
AP000476.1 |
AP000476.1 |
| chr9_+_139839711 | 0.20 |
ENST00000224181.3 |
C8G |
complement component 8, gamma polypeptide |
| chr11_-_61582579 | 0.19 |
ENST00000539419.1 ENST00000545245.1 ENST00000545405.1 ENST00000542506.1 |
FADS1 |
fatty acid desaturase 1 |
| chr1_+_200993071 | 0.19 |
ENST00000446333.1 ENST00000458003.1 |
RP11-168O16.1 |
RP11-168O16.1 |
| chr12_-_63328817 | 0.18 |
ENST00000228705.6 |
PPM1H |
protein phosphatase, Mg2+/Mn2+ dependent, 1H |
| chr6_+_46661575 | 0.18 |
ENST00000450697.1 |
TDRD6 |
tudor domain containing 6 |
| chr4_-_165898768 | 0.18 |
ENST00000329314.5 |
TRIM61 |
tripartite motif containing 61 |
| chr11_+_65266507 | 0.18 |
ENST00000544868.1 |
MALAT1 |
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chr2_+_169926047 | 0.18 |
ENST00000428522.1 ENST00000450153.1 ENST00000421653.1 |
DHRS9 |
dehydrogenase/reductase (SDR family) member 9 |
| chr4_+_6576895 | 0.17 |
ENST00000285599.3 ENST00000504248.1 ENST00000505907.1 |
MAN2B2 |
mannosidase, alpha, class 2B, member 2 |
| chr2_+_233527443 | 0.17 |
ENST00000410095.1 |
EFHD1 |
EF-hand domain family, member D1 |
| chrX_+_72667090 | 0.16 |
ENST00000373514.2 |
CDX4 |
caudal type homeobox 4 |
| chr2_+_210517895 | 0.16 |
ENST00000447185.1 |
MAP2 |
microtubule-associated protein 2 |
| chr11_-_60719213 | 0.16 |
ENST00000227880.3 |
SLC15A3 |
solute carrier family 15 (oligopeptide transporter), member 3 |
| chr10_-_33623826 | 0.15 |
ENST00000374867.2 |
NRP1 |
neuropilin 1 |
| chr9_+_139839686 | 0.15 |
ENST00000371634.2 |
C8G |
complement component 8, gamma polypeptide |
| chr10_+_54074033 | 0.15 |
ENST00000373970.3 |
DKK1 |
dickkopf WNT signaling pathway inhibitor 1 |
| chr17_+_31254892 | 0.14 |
ENST00000394642.3 ENST00000579849.1 |
TMEM98 |
transmembrane protein 98 |
| chr11_-_64527425 | 0.14 |
ENST00000377432.3 |
PYGM |
phosphorylase, glycogen, muscle |
| chr10_-_106240032 | 0.14 |
ENST00000447860.1 |
RP11-127O4.3 |
RP11-127O4.3 |
| chr7_+_100728720 | 0.13 |
ENST00000306085.6 ENST00000412507.1 |
TRIM56 |
tripartite motif containing 56 |
| chr17_-_34756219 | 0.13 |
ENST00000451448.2 ENST00000394359.3 |
TBC1D3C TBC1D3H |
TBC1 domain family, member 3C TBC1 domain family, member 3H |
| chr10_-_134756030 | 0.13 |
ENST00000368586.5 ENST00000368582.2 |
TTC40 |
tetratricopeptide repeat domain 40 |
| chr12_+_79439405 | 0.13 |
ENST00000552744.1 |
SYT1 |
synaptotagmin I |
| chr20_+_3801162 | 0.13 |
ENST00000379573.2 ENST00000379567.2 ENST00000455742.1 ENST00000246041.2 |
AP5S1 |
adaptor-related protein complex 5, sigma 1 subunit |
| chr17_-_36347835 | 0.13 |
ENST00000519532.1 |
TBC1D3 |
TBC1 domain family, member 3 |
| chr7_-_92777606 | 0.13 |
ENST00000437805.1 ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L |
sterile alpha motif domain containing 9-like |
| chr17_-_36348610 | 0.13 |
ENST00000339023.4 ENST00000354664.4 |
TBC1D3 |
TBC1 domain family, member 3 |
| chr17_+_36284791 | 0.12 |
ENST00000505415.1 |
TBC1D3F |
TBC1 domain family, member 3F |
| chr10_-_22292613 | 0.12 |
ENST00000376980.3 |
DNAJC1 |
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr5_+_133861790 | 0.12 |
ENST00000395003.1 |
PHF15 |
jade family PHD finger 2 |
| chr10_-_33624002 | 0.12 |
ENST00000432372.2 |
NRP1 |
neuropilin 1 |
| chr16_+_23765948 | 0.12 |
ENST00000300113.2 |
CHP2 |
calcineurin-like EF-hand protein 2 |
| chr9_+_131799213 | 0.12 |
ENST00000358369.4 ENST00000406926.2 ENST00000277475.5 ENST00000450073.1 |
FAM73B |
family with sequence similarity 73, member B |
| chr17_+_27895609 | 0.12 |
ENST00000581411.2 ENST00000301057.7 |
TP53I13 |
tumor protein p53 inducible protein 13 |
| chr16_-_2260834 | 0.12 |
ENST00000562360.1 ENST00000566018.1 |
BRICD5 |
BRICHOS domain containing 5 |
| chr10_-_13523073 | 0.12 |
ENST00000440282.1 |
BEND7 |
BEN domain containing 7 |
| chr10_-_33623564 | 0.11 |
ENST00000374875.1 ENST00000374822.4 |
NRP1 |
neuropilin 1 |
| chr17_-_34808047 | 0.11 |
ENST00000592614.1 ENST00000591542.1 ENST00000330458.7 ENST00000341264.6 ENST00000592987.1 ENST00000400684.4 |
TBC1D3G TBC1D3H |
TBC1 domain family, member 3G TBC1 domain family, member 3H |
| chr17_-_34807272 | 0.11 |
ENST00000535592.1 ENST00000394453.1 |
TBC1D3G |
TBC1 domain family, member 3G |
| chr19_-_44860820 | 0.11 |
ENST00000354340.4 ENST00000337401.4 ENST00000587909.1 |
ZNF112 |
zinc finger protein 112 |
| chr2_-_32489922 | 0.11 |
ENST00000402280.1 |
NLRC4 |
NLR family, CARD domain containing 4 |
| chr2_-_203736334 | 0.11 |
ENST00000392237.2 ENST00000416760.1 ENST00000412210.1 |
ICA1L |
islet cell autoantigen 1,69kDa-like |
| chr10_-_104001231 | 0.11 |
ENST00000370002.3 |
PITX3 |
paired-like homeodomain 3 |
| chr17_-_64225508 | 0.11 |
ENST00000205948.6 |
APOH |
apolipoprotein H (beta-2-glycoprotein I) |
| chr2_+_46926326 | 0.10 |
ENST00000394861.2 |
SOCS5 |
suppressor of cytokine signaling 5 |
| chr3_-_178969403 | 0.10 |
ENST00000314235.5 ENST00000392685.2 |
KCNMB3 |
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr3_-_156534754 | 0.10 |
ENST00000472943.1 ENST00000473352.1 |
LINC00886 |
long intergenic non-protein coding RNA 886 |
| chr12_-_6579808 | 0.10 |
ENST00000535180.1 ENST00000400911.3 |
VAMP1 |
vesicle-associated membrane protein 1 (synaptobrevin 1) |
| chr16_+_532503 | 0.10 |
ENST00000412256.1 |
RAB11FIP3 |
RAB11 family interacting protein 3 (class II) |
| chr10_-_22292675 | 0.10 |
ENST00000376946.1 |
DNAJC1 |
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr1_-_154474589 | 0.10 |
ENST00000304760.2 |
SHE |
Src homology 2 domain containing E |
| chr8_+_26371763 | 0.10 |
ENST00000521913.1 |
DPYSL2 |
dihydropyrimidinase-like 2 |
| chr3_-_122283079 | 0.09 |
ENST00000471785.1 ENST00000466126.1 |
PARP9 |
poly (ADP-ribose) polymerase family, member 9 |
| chr1_-_99470368 | 0.09 |
ENST00000263177.4 |
LPPR5 |
Lipid phosphate phosphatase-related protein type 5 |
| chr1_-_205325850 | 0.09 |
ENST00000537168.1 |
KLHDC8A |
kelch domain containing 8A |
| chr16_-_67281413 | 0.09 |
ENST00000258201.4 |
FHOD1 |
formin homology 2 domain containing 1 |
| chr9_-_138391692 | 0.09 |
ENST00000429260.2 |
C9orf116 |
chromosome 9 open reading frame 116 |
| chr15_-_42565606 | 0.09 |
ENST00000307216.6 ENST00000448392.1 |
TMEM87A |
transmembrane protein 87A |
| chr7_+_143701022 | 0.09 |
ENST00000408922.2 |
OR6B1 |
olfactory receptor, family 6, subfamily B, member 1 |
| chr1_-_54519134 | 0.09 |
ENST00000371341.1 |
TMEM59 |
transmembrane protein 59 |
| chr11_+_113779704 | 0.09 |
ENST00000537778.1 |
HTR3B |
5-hydroxytryptamine (serotonin) receptor 3B, ionotropic |
| chr6_+_56820018 | 0.09 |
ENST00000370746.3 |
BEND6 |
BEN domain containing 6 |
| chr12_+_52626898 | 0.09 |
ENST00000331817.5 |
KRT7 |
keratin 7 |
| chr1_-_99470558 | 0.09 |
ENST00000370188.3 |
LPPR5 |
Lipid phosphate phosphatase-related protein type 5 |
| chr19_-_12595586 | 0.09 |
ENST00000397732.3 |
ZNF709 |
zinc finger protein 709 |
| chr20_+_39765581 | 0.09 |
ENST00000244007.3 |
PLCG1 |
phospholipase C, gamma 1 |
| chr16_-_67217844 | 0.08 |
ENST00000563902.1 ENST00000561621.1 ENST00000290881.7 |
KIAA0895L |
KIAA0895-like |
| chr8_-_135522425 | 0.08 |
ENST00000521673.1 |
ZFAT |
zinc finger and AT hook domain containing |
| chr11_-_82782861 | 0.08 |
ENST00000524635.1 ENST00000526205.1 ENST00000527633.1 ENST00000533486.1 ENST00000533276.2 |
RAB30 |
RAB30, member RAS oncogene family |
| chr15_+_41952591 | 0.08 |
ENST00000566718.1 ENST00000219905.7 ENST00000389936.4 ENST00000545763.1 |
MGA |
MGA, MAX dimerization protein |
| chr10_+_47894572 | 0.08 |
ENST00000355876.5 |
FAM21B |
family with sequence similarity 21, member B |
| chr11_-_111383064 | 0.08 |
ENST00000525791.1 ENST00000456861.2 ENST00000356018.2 |
BTG4 |
B-cell translocation gene 4 |
| chr7_+_120590803 | 0.08 |
ENST00000315870.5 ENST00000339121.5 ENST00000445699.1 |
ING3 |
inhibitor of growth family, member 3 |
| chr17_-_34591208 | 0.08 |
ENST00000336331.5 |
TBC1D3C |
TBC1 domain family, member 3C |
| chr14_+_22984601 | 0.08 |
ENST00000390509.1 |
TRAJ28 |
T cell receptor alpha joining 28 |
| chr1_-_54519067 | 0.08 |
ENST00000452421.1 ENST00000420738.1 ENST00000234831.5 ENST00000440019.1 |
TMEM59 |
transmembrane protein 59 |
| chr2_-_25194476 | 0.08 |
ENST00000534855.1 |
DNAJC27 |
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr8_-_145028013 | 0.08 |
ENST00000354958.2 |
PLEC |
plectin |
| chr11_-_111794446 | 0.08 |
ENST00000527950.1 |
CRYAB |
crystallin, alpha B |
| chr5_-_179227540 | 0.07 |
ENST00000520875.1 |
MGAT4B |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B |
| chr1_-_221915418 | 0.07 |
ENST00000323825.3 ENST00000366899.3 |
DUSP10 |
dual specificity phosphatase 10 |
| chr16_+_14802801 | 0.07 |
ENST00000526520.1 ENST00000531598.2 |
NPIPA3 |
nuclear pore complex interacting protein family, member A3 |
| chrX_+_153770421 | 0.07 |
ENST00000369609.5 ENST00000369607.1 |
IKBKG |
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr12_-_96390063 | 0.07 |
ENST00000541929.1 |
HAL |
histidine ammonia-lyase |
| chr7_+_29234101 | 0.07 |
ENST00000435288.2 |
CHN2 |
chimerin 2 |
| chr17_-_79791118 | 0.07 |
ENST00000576431.1 ENST00000575061.1 ENST00000455127.2 ENST00000572645.1 ENST00000538396.1 ENST00000573478.1 |
FAM195B |
family with sequence similarity 195, member B |
| chr16_-_31105870 | 0.07 |
ENST00000394971.3 |
VKORC1 |
vitamin K epoxide reductase complex, subunit 1 |
| chr12_+_56552128 | 0.07 |
ENST00000548580.1 ENST00000293422.5 ENST00000348108.4 ENST00000549017.1 ENST00000549566.1 ENST00000536128.1 ENST00000547649.1 ENST00000547408.1 ENST00000551589.1 ENST00000549392.1 ENST00000548400.1 ENST00000548293.1 |
MYL6 |
myosin, light chain 6, alkali, smooth muscle and non-muscle |
| chr16_-_28936493 | 0.07 |
ENST00000544477.1 ENST00000357573.6 |
RABEP2 |
rabaptin, RAB GTPase binding effector protein 2 |
| chr11_-_57177586 | 0.07 |
ENST00000529411.1 |
RP11-872D17.8 |
Uncharacterized protein |
| chr5_-_177210399 | 0.07 |
ENST00000510276.1 |
FAM153A |
family with sequence similarity 153, member A |
| chr20_-_61992738 | 0.06 |
ENST00000370263.4 |
CHRNA4 |
cholinergic receptor, nicotinic, alpha 4 (neuronal) |
| chr16_-_28937027 | 0.06 |
ENST00000358201.4 |
RABEP2 |
rabaptin, RAB GTPase binding effector protein 2 |
| chr17_-_79805146 | 0.06 |
ENST00000415593.1 |
P4HB |
prolyl 4-hydroxylase, beta polypeptide |
| chr17_-_6616678 | 0.06 |
ENST00000381074.4 ENST00000293800.6 ENST00000572352.1 ENST00000576323.1 ENST00000573648.1 |
SLC13A5 |
solute carrier family 13 (sodium-dependent citrate transporter), member 5 |
| chr7_+_29234028 | 0.06 |
ENST00000222792.6 |
CHN2 |
chimerin 2 |
| chr16_+_67207872 | 0.06 |
ENST00000563258.1 ENST00000568146.1 |
NOL3 |
nucleolar protein 3 (apoptosis repressor with CARD domain) |
| chr5_-_218251 | 0.06 |
ENST00000296824.3 |
CCDC127 |
coiled-coil domain containing 127 |
| chr5_+_175487692 | 0.06 |
ENST00000510151.1 |
FAM153B |
family with sequence similarity 153, member B |
| chr16_+_67207838 | 0.06 |
ENST00000566871.1 ENST00000268605.7 |
NOL3 |
nucleolar protein 3 (apoptosis repressor with CARD domain) |
| chr11_+_65154070 | 0.06 |
ENST00000317568.5 ENST00000531296.1 ENST00000533782.1 ENST00000355991.5 ENST00000416776.2 ENST00000526201.1 |
FRMD8 |
FERM domain containing 8 |
| chr22_-_17680472 | 0.06 |
ENST00000330232.4 |
CECR1 |
cat eye syndrome chromosome region, candidate 1 |
| chr4_-_39640700 | 0.06 |
ENST00000295958.5 |
SMIM14 |
small integral membrane protein 14 |
| chr22_+_23213658 | 0.06 |
ENST00000390318.2 |
IGLV4-3 |
immunoglobulin lambda variable 4-3 |
| chr17_-_42992856 | 0.06 |
ENST00000588316.1 ENST00000435360.2 ENST00000586793.1 ENST00000588735.1 ENST00000588037.1 ENST00000592320.1 ENST00000253408.5 |
GFAP |
glial fibrillary acidic protein |
| chr8_+_75512010 | 0.06 |
ENST00000518190.1 ENST00000523118.1 |
RP11-758M4.1 |
Uncharacterized protein |
| chr16_-_31106048 | 0.06 |
ENST00000300851.6 |
VKORC1 |
vitamin K epoxide reductase complex, subunit 1 |
| chr20_-_52790512 | 0.06 |
ENST00000216862.3 |
CYP24A1 |
cytochrome P450, family 24, subfamily A, polypeptide 1 |
| chr16_-_31106211 | 0.06 |
ENST00000532364.1 ENST00000529564.1 ENST00000319788.7 ENST00000354895.4 ENST00000394975.2 |
RP11-196G11.1 VKORC1 |
Uncharacterized protein vitamin K epoxide reductase complex, subunit 1 |
| chr14_-_24806588 | 0.06 |
ENST00000555591.1 ENST00000554569.1 |
RP11-934B9.3 RIPK3 |
Uncharacterized protein receptor-interacting serine-threonine kinase 3 |
| chr10_-_18940501 | 0.06 |
ENST00000377304.4 |
NSUN6 |
NOP2/Sun domain family, member 6 |
| chr5_-_145562147 | 0.06 |
ENST00000545646.1 ENST00000274562.9 ENST00000510191.1 ENST00000394434.2 |
LARS |
leucyl-tRNA synthetase |
| chr1_+_47137445 | 0.06 |
ENST00000569393.1 ENST00000334122.4 ENST00000415500.1 |
TEX38 |
testis expressed 38 |
| chr3_+_46618727 | 0.06 |
ENST00000296145.5 |
TDGF1 |
teratocarcinoma-derived growth factor 1 |
| chr7_+_94536898 | 0.06 |
ENST00000433360.1 ENST00000340694.4 ENST00000424654.1 |
PPP1R9A |
protein phosphatase 1, regulatory subunit 9A |
| chr16_-_2770216 | 0.06 |
ENST00000302641.3 |
PRSS27 |
protease, serine 27 |
| chr11_-_82782952 | 0.06 |
ENST00000534141.1 |
RAB30 |
RAB30, member RAS oncogene family |
| chr1_+_3816936 | 0.06 |
ENST00000413332.1 ENST00000442673.1 ENST00000439488.1 |
RP13-15E13.1 |
long intergenic non-protein coding RNA 1134 |
| chr2_-_230786679 | 0.06 |
ENST00000543084.1 ENST00000343290.5 ENST00000389044.4 ENST00000283943.5 |
TRIP12 |
thyroid hormone receptor interactor 12 |
| chr8_+_24298531 | 0.06 |
ENST00000175238.6 |
ADAM7 |
ADAM metallopeptidase domain 7 |
| chr3_+_2140565 | 0.06 |
ENST00000455083.1 ENST00000418658.1 |
CNTN4 |
contactin 4 |
| chr19_-_19932501 | 0.06 |
ENST00000540806.2 ENST00000590766.1 ENST00000587452.1 ENST00000545006.1 ENST00000590319.1 ENST00000587461.1 ENST00000450683.2 ENST00000443905.2 ENST00000590274.1 |
ZNF506 CTC-559E9.4 |
zinc finger protein 506 CTC-559E9.4 |
| chr13_+_24844819 | 0.06 |
ENST00000399949.2 |
SPATA13 |
spermatogenesis associated 13 |
| chr8_+_24298597 | 0.05 |
ENST00000380789.1 |
ADAM7 |
ADAM metallopeptidase domain 7 |
| chr3_-_3151664 | 0.05 |
ENST00000256452.3 ENST00000311981.8 ENST00000430514.2 ENST00000456302.1 |
IL5RA |
interleukin 5 receptor, alpha |
| chr1_+_26348259 | 0.05 |
ENST00000374280.3 |
EXTL1 |
exostosin-like glycosyltransferase 1 |
| chr1_+_109234907 | 0.05 |
ENST00000370025.4 ENST00000370022.5 ENST00000370021.1 |
PRPF38B |
pre-mRNA processing factor 38B |
| chr2_+_108905325 | 0.05 |
ENST00000438339.1 ENST00000409880.1 ENST00000437390.2 |
SULT1C2 |
sulfotransferase family, cytosolic, 1C, member 2 |
| chr17_-_79212825 | 0.05 |
ENST00000374769.2 |
ENTHD2 |
ENTH domain containing 2 |
| chr3_-_54962100 | 0.05 |
ENST00000273286.5 |
LRTM1 |
leucine-rich repeats and transmembrane domains 1 |
| chr6_+_167704798 | 0.05 |
ENST00000230256.3 |
UNC93A |
unc-93 homolog A (C. elegans) |
| chr14_-_54908043 | 0.05 |
ENST00000556113.1 ENST00000553660.1 ENST00000395573.4 ENST00000557690.1 ENST00000216416.4 |
CNIH1 |
cornichon family AMPA receptor auxiliary protein 1 |
| chr10_+_119301928 | 0.05 |
ENST00000553456.3 |
EMX2 |
empty spiracles homeobox 2 |
| chr2_+_176957619 | 0.05 |
ENST00000392539.3 |
HOXD13 |
homeobox D13 |
| chr5_+_177433973 | 0.05 |
ENST00000507848.1 |
FAM153C |
family with sequence similarity 153, member C |
| chr12_-_96390108 | 0.05 |
ENST00000538703.1 ENST00000261208.3 |
HAL |
histidine ammonia-lyase |
| chr22_+_29664305 | 0.05 |
ENST00000414183.2 ENST00000333395.6 ENST00000455726.1 ENST00000332035.6 |
EWSR1 |
EWS RNA-binding protein 1 |
| chrX_+_43515467 | 0.05 |
ENST00000338702.3 ENST00000542639.1 |
MAOA |
monoamine oxidase A |
| chr8_+_10383039 | 0.05 |
ENST00000328655.3 ENST00000522210.1 |
PRSS55 |
protease, serine, 55 |
| chr15_+_83776324 | 0.05 |
ENST00000379390.6 ENST00000379386.4 ENST00000565774.1 ENST00000565982.1 |
TM6SF1 |
transmembrane 6 superfamily member 1 |
| chr9_-_139343294 | 0.05 |
ENST00000313084.5 |
SEC16A |
SEC16 homolog A (S. cerevisiae) |
| chr11_-_73882029 | 0.05 |
ENST00000539061.1 |
C2CD3 |
C2 calcium-dependent domain containing 3 |
| chr9_-_15307200 | 0.05 |
ENST00000506891.1 ENST00000541445.1 ENST00000512701.2 ENST00000380850.4 ENST00000297615.5 ENST00000355694.2 |
TTC39B |
tetratricopeptide repeat domain 39B |
| chr11_+_6624955 | 0.05 |
ENST00000299421.4 ENST00000537806.1 |
ILK |
integrin-linked kinase |
| chr11_+_6625046 | 0.05 |
ENST00000396751.2 |
ILK |
integrin-linked kinase |
| chr5_+_140560980 | 0.05 |
ENST00000361016.2 |
PCDHB16 |
protocadherin beta 16 |
| chr4_+_142557771 | 0.05 |
ENST00000514653.1 |
IL15 |
interleukin 15 |
| chr17_+_43224684 | 0.05 |
ENST00000332499.2 |
HEXIM1 |
hexamethylene bis-acetamide inducible 1 |
| chr11_-_73882057 | 0.05 |
ENST00000334126.7 ENST00000313663.7 |
C2CD3 |
C2 calcium-dependent domain containing 3 |
| chr11_+_6624970 | 0.05 |
ENST00000420936.2 ENST00000528995.1 |
ILK |
integrin-linked kinase |
| chr11_-_506316 | 0.05 |
ENST00000532055.1 ENST00000531540.1 |
RNH1 |
ribonuclease/angiogenin inhibitor 1 |
| chr3_-_93781750 | 0.05 |
ENST00000314636.2 |
DHFRL1 |
dihydrofolate reductase-like 1 |
| chr1_+_52082751 | 0.05 |
ENST00000447887.1 ENST00000435686.2 ENST00000428468.1 ENST00000453295.1 |
OSBPL9 |
oxysterol binding protein-like 9 |
| chr11_+_64358686 | 0.05 |
ENST00000473690.1 |
SLC22A12 |
solute carrier family 22 (organic anion/urate transporter), member 12 |
| chr4_-_103266219 | 0.05 |
ENST00000394833.2 |
SLC39A8 |
solute carrier family 39 (zinc transporter), member 8 |
| chrX_-_130423200 | 0.04 |
ENST00000361420.3 |
IGSF1 |
immunoglobulin superfamily, member 1 |
| chr6_+_56819895 | 0.04 |
ENST00000370748.3 |
BEND6 |
BEN domain containing 6 |
| chr6_-_36953833 | 0.04 |
ENST00000538808.1 ENST00000460219.1 ENST00000373616.5 ENST00000373627.5 |
MTCH1 |
mitochondrial carrier 1 |
| chr14_-_101295407 | 0.04 |
ENST00000596284.1 |
AL117190.2 |
AL117190.2 |
| chr13_+_114321463 | 0.04 |
ENST00000335678.6 |
GRK1 |
G protein-coupled receptor kinase 1 |
| chr15_-_81282133 | 0.04 |
ENST00000261758.4 |
MESDC2 |
mesoderm development candidate 2 |
| chrX_-_13835147 | 0.04 |
ENST00000493677.1 ENST00000355135.2 |
GPM6B |
glycoprotein M6B |
| chr3_+_14693247 | 0.04 |
ENST00000383794.3 ENST00000303688.7 |
CCDC174 |
coiled-coil domain containing 174 |
| chr6_+_31802685 | 0.04 |
ENST00000375639.2 ENST00000375638.3 ENST00000375635.2 ENST00000375642.2 ENST00000395789.1 |
C6orf48 |
chromosome 6 open reading frame 48 |
| chr11_+_82783097 | 0.04 |
ENST00000501011.2 ENST00000527627.1 ENST00000526795.1 ENST00000533528.1 ENST00000533708.1 ENST00000534499.1 |
RAB30-AS1 |
RAB30 antisense RNA 1 (head to head) |
| chr19_+_13261216 | 0.04 |
ENST00000587885.1 ENST00000292433.3 |
IER2 |
immediate early response 2 |
| chr20_+_2276639 | 0.04 |
ENST00000381458.5 |
TGM3 |
transglutaminase 3 |
| chr5_+_175488258 | 0.04 |
ENST00000512862.1 |
FAM153B |
family with sequence similarity 153, member B |
| chr6_+_167704838 | 0.04 |
ENST00000366829.2 |
UNC93A |
unc-93 homolog A (C. elegans) |
| chr5_-_7869108 | 0.04 |
ENST00000264669.5 ENST00000507572.1 ENST00000504695.1 |
FASTKD3 |
FAST kinase domains 3 |
| chr1_+_92414952 | 0.04 |
ENST00000449584.1 ENST00000427104.1 ENST00000355011.3 ENST00000448194.1 ENST00000426141.1 ENST00000450792.1 ENST00000548992.1 ENST00000552654.1 ENST00000457265.1 |
BRDT |
bromodomain, testis-specific |
| chr15_+_41851211 | 0.04 |
ENST00000263798.3 |
TYRO3 |
TYRO3 protein tyrosine kinase |
| chr6_+_30525051 | 0.04 |
ENST00000376557.3 |
PRR3 |
proline rich 3 |
| chr8_-_77912431 | 0.04 |
ENST00000357039.4 ENST00000522527.1 |
PEX2 |
peroxisomal biogenesis factor 2 |
| chr20_+_62612470 | 0.04 |
ENST00000266079.4 ENST00000535781.1 |
PRPF6 |
pre-mRNA processing factor 6 |
| chr14_+_21498360 | 0.04 |
ENST00000321760.6 ENST00000460647.2 ENST00000530140.2 ENST00000472458.1 |
TPPP2 |
tubulin polymerization-promoting protein family member 2 |
| chr14_-_106781017 | 0.04 |
ENST00000390612.2 |
IGHV4-28 |
immunoglobulin heavy variable 4-28 |
| chrX_+_153813407 | 0.04 |
ENST00000443287.2 ENST00000333128.3 |
CTAG1A |
cancer/testis antigen 1A |
| chr9_+_140125209 | 0.04 |
ENST00000538474.1 |
SLC34A3 |
solute carrier family 34 (type II sodium/phosphate contransporter), member 3 |
| chr3_+_111578131 | 0.04 |
ENST00000498699.1 |
PHLDB2 |
pleckstrin homology-like domain, family B, member 2 |
| chr22_-_29663954 | 0.04 |
ENST00000216085.7 |
RHBDD3 |
rhomboid domain containing 3 |
| chr5_-_177209832 | 0.04 |
ENST00000393518.3 ENST00000505531.1 ENST00000503567.1 |
FAM153A |
family with sequence similarity 153, member A |
| chr8_+_119294456 | 0.04 |
ENST00000366457.2 |
AC023590.1 |
Uncharacterized protein |
| chrX_+_150732094 | 0.04 |
ENST00000370357.4 |
PASD1 |
PAS domain containing 1 |
| chr17_-_79212884 | 0.03 |
ENST00000300714.3 |
ENTHD2 |
ENTH domain containing 2 |
| chr4_-_164534657 | 0.03 |
ENST00000339875.5 |
MARCH1 |
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr7_+_99699280 | 0.03 |
ENST00000421755.1 |
AP4M1 |
adaptor-related protein complex 4, mu 1 subunit |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0032571 | response to vitamin K(GO:0032571) bone regeneration(GO:1990523) |
| 0.1 | 0.5 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.1 | 0.4 | GO:0021649 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.1 | 0.4 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.1 | 0.2 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.0 | 0.1 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.0 | 0.3 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.1 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 0.1 | GO:0014876 | response to injury involved in regulation of muscle adaptation(GO:0014876) |
| 0.0 | 0.1 | GO:1904933 | regulation of cell proliferation in midbrain(GO:1904933) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.5 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.0 | 0.1 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.0 | 0.1 | GO:2000452 | CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:0035698) positive regulation of necroptotic process(GO:0060545) regulation of CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:2000452) |
| 0.0 | 0.1 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.0 | 0.1 | GO:1904048 | regulation of spontaneous neurotransmitter secretion(GO:1904048) |
| 0.0 | 0.1 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 0.0 | 0.1 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.0 | 0.1 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.3 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.1 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) regulation of respiratory burst involved in inflammatory response(GO:0060264) |
| 0.0 | 0.2 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.0 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.0 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 0.0 | 0.0 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.2 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.0 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.1 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.2 | GO:0035791 | platelet-derived growth factor receptor-beta signaling pathway(GO:0035791) |
| 0.0 | 0.1 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.1 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.5 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.2 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.4 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.2 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.1 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.0 | GO:0072563 | endothelial microparticle(GO:0072563) |
| 0.0 | 0.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.6 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.2 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 0.4 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.2 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.2 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.0 | 0.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.2 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.0 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.1 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.0 | 0.1 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.0 | 0.2 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.4 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.0 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.0 | 0.1 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.0 | 0.1 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.0 | GO:0015198 | oligopeptide transporter activity(GO:0015198) cobalamin transporter activity(GO:0015235) oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.1 | GO:0017153 | citrate transmembrane transporter activity(GO:0015137) succinate transmembrane transporter activity(GO:0015141) tricarboxylic acid transmembrane transporter activity(GO:0015142) sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.2 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.5 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.0 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.4 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |