Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
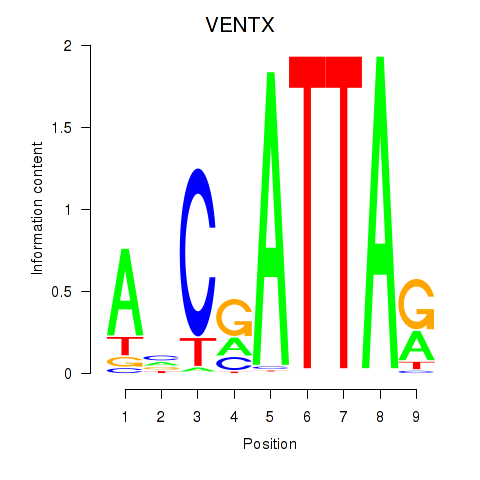
Results for VENTX
Z-value: 0.33
Transcription factors associated with VENTX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
VENTX
|
ENSG00000151650.7 | VENTX |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| VENTX | hg19_v2_chr10_+_135050908_135050908 | -0.19 | 6.5e-01 | Click! |
Activity profile of VENTX motif
Sorted Z-values of VENTX motif
Network of associatons between targets according to the STRING database.
First level regulatory network of VENTX
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_107712173 | 0.39 |
ENST00000280758.5 ENST00000420571.2 |
BTBD11 |
BTB (POZ) domain containing 11 |
| chr14_+_65171315 | 0.31 |
ENST00000394691.1 |
PLEKHG3 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr4_-_155511887 | 0.30 |
ENST00000302053.3 ENST00000403106.3 |
FGA |
fibrinogen alpha chain |
| chr9_-_23825956 | 0.27 |
ENST00000397312.2 |
ELAVL2 |
ELAV like neuron-specific RNA binding protein 2 |
| chr1_+_62439037 | 0.25 |
ENST00000545929.1 |
INADL |
InaD-like (Drosophila) |
| chr12_-_52828147 | 0.24 |
ENST00000252245.5 |
KRT75 |
keratin 75 |
| chr1_+_62417957 | 0.23 |
ENST00000307297.7 ENST00000543708.1 |
INADL |
InaD-like (Drosophila) |
| chr18_+_55888767 | 0.22 |
ENST00000431212.2 ENST00000586268.1 ENST00000587190.1 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chrX_+_105937068 | 0.22 |
ENST00000324342.3 |
RNF128 |
ring finger protein 128, E3 ubiquitin protein ligase |
| chr6_+_130339710 | 0.22 |
ENST00000526087.1 ENST00000533560.1 ENST00000361794.2 |
L3MBTL3 |
l(3)mbt-like 3 (Drosophila) |
| chr5_-_82969405 | 0.22 |
ENST00000510978.1 |
HAPLN1 |
hyaluronan and proteoglycan link protein 1 |
| chr7_-_22234381 | 0.20 |
ENST00000458533.1 |
RAPGEF5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr12_-_12674032 | 0.19 |
ENST00000298573.4 |
DUSP16 |
dual specificity phosphatase 16 |
| chr8_+_107738343 | 0.18 |
ENST00000521592.1 |
OXR1 |
oxidation resistance 1 |
| chr18_-_24237339 | 0.17 |
ENST00000580191.1 |
KCTD1 |
potassium channel tetramerization domain containing 1 |
| chr4_+_71458012 | 0.17 |
ENST00000449493.2 |
AMBN |
ameloblastin (enamel matrix protein) |
| chr2_-_165698521 | 0.16 |
ENST00000409184.3 ENST00000392717.2 ENST00000456693.1 |
COBLL1 |
cordon-bleu WH2 repeat protein-like 1 |
| chr12_+_56473939 | 0.16 |
ENST00000450146.2 |
ERBB3 |
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr8_+_107738240 | 0.16 |
ENST00000449762.2 ENST00000297447.6 |
OXR1 |
oxidation resistance 1 |
| chr20_-_50722183 | 0.15 |
ENST00000371523.4 |
ZFP64 |
ZFP64 zinc finger protein |
| chr10_+_24738355 | 0.15 |
ENST00000307544.6 |
KIAA1217 |
KIAA1217 |
| chr3_-_27764190 | 0.14 |
ENST00000537516.1 |
EOMES |
eomesodermin |
| chr2_-_165698662 | 0.14 |
ENST00000194871.6 ENST00000445474.2 |
COBLL1 |
cordon-bleu WH2 repeat protein-like 1 |
| chr18_-_74207146 | 0.14 |
ENST00000443185.2 |
ZNF516 |
zinc finger protein 516 |
| chr14_+_65171099 | 0.14 |
ENST00000247226.7 |
PLEKHG3 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr4_-_84035868 | 0.13 |
ENST00000426923.2 ENST00000509973.1 |
PLAC8 |
placenta-specific 8 |
| chr8_+_31496809 | 0.13 |
ENST00000518104.1 ENST00000519301.1 |
NRG1 |
neuregulin 1 |
| chr12_-_86650045 | 0.13 |
ENST00000604798.1 |
MGAT4C |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr6_-_66417107 | 0.13 |
ENST00000370621.3 ENST00000370618.3 ENST00000503581.1 ENST00000393380.2 |
EYS |
eyes shut homolog (Drosophila) |
| chr2_+_202937972 | 0.13 |
ENST00000541917.1 ENST00000295844.3 |
AC079354.1 |
uncharacterized protein KIAA2012 |
| chr1_-_23886285 | 0.13 |
ENST00000374561.5 |
ID3 |
inhibitor of DNA binding 3, dominant negative helix-loop-helix protein |
| chr11_+_22359562 | 0.12 |
ENST00000263160.3 |
SLC17A6 |
solute carrier family 17 (vesicular glutamate transporter), member 6 |
| chr12_-_89746173 | 0.12 |
ENST00000308385.6 |
DUSP6 |
dual specificity phosphatase 6 |
| chr16_-_67517716 | 0.12 |
ENST00000290953.2 |
AGRP |
agouti related protein homolog (mouse) |
| chr21_+_39644395 | 0.11 |
ENST00000398934.1 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr1_+_174417095 | 0.11 |
ENST00000367685.2 |
GPR52 |
G protein-coupled receptor 52 |
| chr12_-_57328187 | 0.11 |
ENST00000293502.1 |
SDR9C7 |
short chain dehydrogenase/reductase family 9C, member 7 |
| chr1_+_153747746 | 0.10 |
ENST00000368661.3 |
SLC27A3 |
solute carrier family 27 (fatty acid transporter), member 3 |
| chr6_-_138820624 | 0.10 |
ENST00000343505.5 |
NHSL1 |
NHS-like 1 |
| chr6_+_12290586 | 0.09 |
ENST00000379375.5 |
EDN1 |
endothelin 1 |
| chr4_+_86525299 | 0.09 |
ENST00000512201.1 |
ARHGAP24 |
Rho GTPase activating protein 24 |
| chr1_+_153746683 | 0.09 |
ENST00000271857.2 |
SLC27A3 |
solute carrier family 27 (fatty acid transporter), member 3 |
| chr5_-_156486120 | 0.09 |
ENST00000522693.1 |
HAVCR1 |
hepatitis A virus cellular receptor 1 |
| chr12_-_16758059 | 0.09 |
ENST00000261169.6 |
LMO3 |
LIM domain only 3 (rhombotin-like 2) |
| chr6_-_111927062 | 0.09 |
ENST00000359831.4 |
TRAF3IP2 |
TRAF3 interacting protein 2 |
| chr10_+_114043493 | 0.09 |
ENST00000369422.3 |
TECTB |
tectorin beta |
| chr17_+_17685422 | 0.09 |
ENST00000395774.1 |
RAI1 |
retinoic acid induced 1 |
| chr6_-_32157947 | 0.09 |
ENST00000375050.4 |
PBX2 |
pre-B-cell leukemia homeobox 2 |
| chr2_-_203776864 | 0.09 |
ENST00000261015.4 |
WDR12 |
WD repeat domain 12 |
| chr11_+_35198118 | 0.09 |
ENST00000525211.1 ENST00000526000.1 ENST00000279452.6 ENST00000527889.1 |
CD44 |
CD44 molecule (Indian blood group) |
| chr12_+_81110684 | 0.08 |
ENST00000228644.3 |
MYF5 |
myogenic factor 5 |
| chr11_-_72070206 | 0.08 |
ENST00000544382.1 |
CLPB |
ClpB caseinolytic peptidase B homolog (E. coli) |
| chr8_+_50824233 | 0.08 |
ENST00000522124.1 |
SNTG1 |
syntrophin, gamma 1 |
| chr18_-_53253323 | 0.08 |
ENST00000540999.1 ENST00000563888.2 |
TCF4 |
transcription factor 4 |
| chr13_-_28545276 | 0.08 |
ENST00000381020.7 |
CDX2 |
caudal type homeobox 2 |
| chr4_-_84035905 | 0.07 |
ENST00000311507.4 |
PLAC8 |
placenta-specific 8 |
| chr8_+_26150628 | 0.07 |
ENST00000523925.1 ENST00000315985.7 |
PPP2R2A |
protein phosphatase 2, regulatory subunit B, alpha |
| chrX_+_130192318 | 0.07 |
ENST00000370922.1 |
ARHGAP36 |
Rho GTPase activating protein 36 |
| chr2_+_159825143 | 0.06 |
ENST00000454300.1 ENST00000263635.6 |
TANC1 |
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chr4_-_26492076 | 0.06 |
ENST00000295589.3 |
CCKAR |
cholecystokinin A receptor |
| chr18_-_53253112 | 0.06 |
ENST00000568673.1 ENST00000562847.1 ENST00000568147.1 |
TCF4 |
transcription factor 4 |
| chrX_+_130192216 | 0.06 |
ENST00000276211.5 |
ARHGAP36 |
Rho GTPase activating protein 36 |
| chr11_-_107729887 | 0.06 |
ENST00000525815.1 |
SLC35F2 |
solute carrier family 35, member F2 |
| chrX_-_130423200 | 0.06 |
ENST00000361420.3 |
IGSF1 |
immunoglobulin superfamily, member 1 |
| chr22_+_39101728 | 0.06 |
ENST00000216044.5 ENST00000484657.1 |
GTPBP1 |
GTP binding protein 1 |
| chr12_+_70219052 | 0.06 |
ENST00000552032.2 ENST00000547771.2 |
MYRFL |
myelin regulatory factor-like |
| chr12_-_53012343 | 0.06 |
ENST00000305748.3 |
KRT73 |
keratin 73 |
| chrX_-_46187069 | 0.05 |
ENST00000446884.1 |
RP1-30G7.2 |
RP1-30G7.2 |
| chr13_-_26795840 | 0.05 |
ENST00000381570.3 ENST00000399762.2 ENST00000346166.3 |
RNF6 |
ring finger protein (C3H2C3 type) 6 |
| chr11_-_124190184 | 0.05 |
ENST00000357438.2 |
OR8D2 |
olfactory receptor, family 8, subfamily D, member 2 |
| chr1_+_244515930 | 0.05 |
ENST00000366537.1 ENST00000308105.4 |
C1orf100 |
chromosome 1 open reading frame 100 |
| chr6_-_111927449 | 0.05 |
ENST00000368761.5 ENST00000392556.4 ENST00000340026.6 |
TRAF3IP2 |
TRAF3 interacting protein 2 |
| chr1_-_178840157 | 0.05 |
ENST00000367629.1 ENST00000234816.2 |
ANGPTL1 |
angiopoietin-like 1 |
| chr7_-_104909435 | 0.05 |
ENST00000357311.3 |
SRPK2 |
SRSF protein kinase 2 |
| chr8_-_87755878 | 0.05 |
ENST00000320005.5 |
CNGB3 |
cyclic nucleotide gated channel beta 3 |
| chr17_-_48785216 | 0.05 |
ENST00000285243.6 |
ANKRD40 |
ankyrin repeat domain 40 |
| chr14_+_32798547 | 0.05 |
ENST00000557354.1 ENST00000557102.1 ENST00000557272.1 |
AKAP6 |
A kinase (PRKA) anchor protein 6 |
| chr17_-_46716647 | 0.05 |
ENST00000608940.1 |
RP11-357H14.17 |
RP11-357H14.17 |
| chr4_+_169013666 | 0.05 |
ENST00000359299.3 |
ANXA10 |
annexin A10 |
| chr6_+_152011628 | 0.05 |
ENST00000404742.1 ENST00000440973.1 |
ESR1 |
estrogen receptor 1 |
| chr2_-_163008903 | 0.05 |
ENST00000418842.2 ENST00000375497.3 |
GCG |
glucagon |
| chr10_-_62332357 | 0.05 |
ENST00000503366.1 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
| chr9_+_116225999 | 0.05 |
ENST00000317613.6 |
RGS3 |
regulator of G-protein signaling 3 |
| chr18_+_59000815 | 0.05 |
ENST00000262717.4 |
CDH20 |
cadherin 20, type 2 |
| chr11_-_11374904 | 0.04 |
ENST00000528848.2 |
CSNK2A3 |
casein kinase 2, alpha 3 polypeptide |
| chr18_-_53804580 | 0.04 |
ENST00000590484.1 ENST00000589293.1 ENST00000587904.1 ENST00000591974.1 |
RP11-456O19.4 |
RP11-456O19.4 |
| chr19_-_49622348 | 0.04 |
ENST00000408991.2 |
C19orf73 |
chromosome 19 open reading frame 73 |
| chr17_-_7307358 | 0.04 |
ENST00000576017.1 ENST00000302422.3 ENST00000535512.1 |
TMEM256 TMEM256-PLSCR3 |
transmembrane protein 256 TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr12_+_25205446 | 0.04 |
ENST00000557489.1 ENST00000354454.3 ENST00000536173.1 |
LRMP |
lymphoid-restricted membrane protein |
| chr12_-_101604185 | 0.04 |
ENST00000536262.2 |
SLC5A8 |
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 8 |
| chrX_-_139587225 | 0.04 |
ENST00000370536.2 |
SOX3 |
SRY (sex determining region Y)-box 3 |
| chr8_-_103425047 | 0.04 |
ENST00000520539.1 |
UBR5 |
ubiquitin protein ligase E3 component n-recognin 5 |
| chr5_+_61874562 | 0.04 |
ENST00000334994.5 ENST00000409534.1 |
LRRC70 IPO11 |
leucine rich repeat containing 70 importin 11 |
| chr6_+_37897735 | 0.04 |
ENST00000373389.5 |
ZFAND3 |
zinc finger, AN1-type domain 3 |
| chrX_+_78003204 | 0.04 |
ENST00000435339.3 ENST00000514744.1 |
LPAR4 |
lysophosphatidic acid receptor 4 |
| chr17_+_43239191 | 0.04 |
ENST00000589230.1 |
HEXIM2 |
hexamethylene bis-acetamide inducible 2 |
| chr5_+_1225470 | 0.04 |
ENST00000324642.3 ENST00000296821.4 |
SLC6A18 |
solute carrier family 6 (neutral amino acid transporter), member 18 |
| chr12_-_122018346 | 0.04 |
ENST00000377069.4 |
KDM2B |
lysine (K)-specific demethylase 2B |
| chr9_+_117904097 | 0.04 |
ENST00000374016.1 |
DEC1 |
deleted in esophageal cancer 1 |
| chr17_-_10372875 | 0.04 |
ENST00000255381.2 |
MYH4 |
myosin, heavy chain 4, skeletal muscle |
| chr5_+_60933634 | 0.04 |
ENST00000505642.1 |
C5orf64 |
chromosome 5 open reading frame 64 |
| chr10_+_74451883 | 0.04 |
ENST00000373053.3 ENST00000357157.6 |
MCU |
mitochondrial calcium uniporter |
| chr6_-_139613269 | 0.04 |
ENST00000358430.3 |
TXLNB |
taxilin beta |
| chr8_-_103424916 | 0.04 |
ENST00000220959.4 |
UBR5 |
ubiquitin protein ligase E3 component n-recognin 5 |
| chr6_-_39693111 | 0.04 |
ENST00000373215.3 ENST00000538893.1 ENST00000287152.7 ENST00000373216.3 |
KIF6 |
kinesin family member 6 |
| chr1_+_150337100 | 0.04 |
ENST00000401000.4 |
RPRD2 |
regulation of nuclear pre-mRNA domain containing 2 |
| chr4_+_119200215 | 0.04 |
ENST00000602573.1 |
SNHG8 |
small nucleolar RNA host gene 8 (non-protein coding) |
| chr3_-_72149553 | 0.03 |
ENST00000468646.2 ENST00000464271.1 |
LINC00877 |
long intergenic non-protein coding RNA 877 |
| chr14_+_22670455 | 0.03 |
ENST00000390460.1 |
TRAV26-2 |
T cell receptor alpha variable 26-2 |
| chr3_+_156799587 | 0.03 |
ENST00000469196.1 |
RP11-6F2.5 |
RP11-6F2.5 |
| chr14_-_20801427 | 0.03 |
ENST00000557665.1 ENST00000358932.4 ENST00000353689.4 |
CCNB1IP1 |
cyclin B1 interacting protein 1, E3 ubiquitin protein ligase |
| chr8_-_103424986 | 0.03 |
ENST00000521922.1 |
UBR5 |
ubiquitin protein ligase E3 component n-recognin 5 |
| chr14_+_32798462 | 0.03 |
ENST00000280979.4 |
AKAP6 |
A kinase (PRKA) anchor protein 6 |
| chr2_+_171034646 | 0.03 |
ENST00000409044.3 ENST00000408978.4 |
MYO3B |
myosin IIIB |
| chr20_-_49307897 | 0.03 |
ENST00000535356.1 |
FAM65C |
family with sequence similarity 65, member C |
| chr11_+_64018955 | 0.03 |
ENST00000279230.6 ENST00000540288.1 ENST00000325234.5 |
PLCB3 |
phospholipase C, beta 3 (phosphatidylinositol-specific) |
| chr9_+_136501478 | 0.03 |
ENST00000393056.2 ENST00000263611.2 |
DBH |
dopamine beta-hydroxylase (dopamine beta-monooxygenase) |
| chr3_+_171561127 | 0.03 |
ENST00000334567.5 ENST00000450693.1 |
TMEM212 |
transmembrane protein 212 |
| chr8_-_109799793 | 0.03 |
ENST00000297459.3 |
TMEM74 |
transmembrane protein 74 |
| chr7_+_129984630 | 0.03 |
ENST00000355388.3 ENST00000497503.1 ENST00000463587.1 ENST00000461828.1 ENST00000494311.1 ENST00000466363.2 ENST00000485477.1 ENST00000431780.2 ENST00000474905.1 |
CPA5 |
carboxypeptidase A5 |
| chr1_+_41174988 | 0.03 |
ENST00000372652.1 |
NFYC |
nuclear transcription factor Y, gamma |
| chr20_-_49308048 | 0.03 |
ENST00000327979.2 |
FAM65C |
family with sequence similarity 65, member C |
| chr11_+_75526212 | 0.03 |
ENST00000356136.3 |
UVRAG |
UV radiation resistance associated |
| chr1_+_150337144 | 0.03 |
ENST00000539519.1 ENST00000369067.3 ENST00000369068.4 |
RPRD2 |
regulation of nuclear pre-mRNA domain containing 2 |
| chr17_+_68100989 | 0.03 |
ENST00000585558.1 ENST00000392670.1 |
KCNJ16 |
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr11_+_46722368 | 0.02 |
ENST00000311764.2 |
ZNF408 |
zinc finger protein 408 |
| chr4_+_130017268 | 0.02 |
ENST00000425929.1 ENST00000508673.1 ENST00000508622.1 |
C4orf33 |
chromosome 4 open reading frame 33 |
| chr14_+_22309368 | 0.02 |
ENST00000390433.1 |
TRAV12-1 |
T cell receptor alpha variable 12-1 |
| chr3_+_185303962 | 0.02 |
ENST00000296257.5 |
SENP2 |
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chr2_+_45168875 | 0.02 |
ENST00000260653.3 |
SIX3 |
SIX homeobox 3 |
| chr3_+_32859510 | 0.02 |
ENST00000383763.5 |
TRIM71 |
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr3_+_130569429 | 0.02 |
ENST00000505330.1 ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1 |
ATPase, Ca++ transporting, type 2C, member 1 |
| chr19_+_48949030 | 0.02 |
ENST00000253237.5 |
GRWD1 |
glutamate-rich WD repeat containing 1 |
| chr6_-_76203345 | 0.01 |
ENST00000393004.2 |
FILIP1 |
filamin A interacting protein 1 |
| chr10_-_56561022 | 0.01 |
ENST00000373965.2 ENST00000414778.1 ENST00000395438.1 ENST00000409834.1 ENST00000395445.1 ENST00000395446.1 ENST00000395442.1 ENST00000395440.1 ENST00000395432.2 ENST00000361849.3 ENST00000395433.1 ENST00000320301.6 ENST00000395430.1 ENST00000437009.1 |
PCDH15 |
protocadherin-related 15 |
| chr11_-_64684672 | 0.01 |
ENST00000377264.3 ENST00000421419.2 |
ATG2A |
autophagy related 2A |
| chr17_-_47045949 | 0.01 |
ENST00000357424.2 |
GIP |
gastric inhibitory polypeptide |
| chr2_+_166152283 | 0.01 |
ENST00000375427.2 |
SCN2A |
sodium channel, voltage-gated, type II, alpha subunit |
| chr6_+_140175987 | 0.01 |
ENST00000414038.1 ENST00000431609.1 |
RP5-899B16.1 |
RP5-899B16.1 |
| chr17_-_202579 | 0.01 |
ENST00000577079.1 ENST00000331302.7 ENST00000536489.2 |
RPH3AL |
rabphilin 3A-like (without C2 domains) |
| chr12_+_8666126 | 0.01 |
ENST00000299665.2 |
CLEC4D |
C-type lectin domain family 4, member D |
| chr10_-_56560939 | 0.01 |
ENST00000373955.1 |
PCDH15 |
protocadherin-related 15 |
| chr4_+_86748898 | 0.01 |
ENST00000509300.1 |
ARHGAP24 |
Rho GTPase activating protein 24 |
| chr1_-_190446759 | 0.01 |
ENST00000367462.3 |
BRINP3 |
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr4_-_46126093 | 0.01 |
ENST00000295452.4 |
GABRG1 |
gamma-aminobutyric acid (GABA) A receptor, gamma 1 |
| chr20_+_42136308 | 0.01 |
ENST00000434666.1 ENST00000427442.2 ENST00000439769.1 ENST00000418998.1 |
L3MBTL1 |
l(3)mbt-like 1 (Drosophila) |
| chr2_+_226265364 | 0.01 |
ENST00000272907.6 |
NYAP2 |
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2 |
| chr12_+_25205568 | 0.01 |
ENST00000548766.1 ENST00000556887.1 |
LRMP |
lymphoid-restricted membrane protein |
| chrX_+_106163626 | 0.01 |
ENST00000336803.1 |
CLDN2 |
claudin 2 |
| chr4_-_168155169 | 0.01 |
ENST00000534949.1 ENST00000535728.1 |
SPOCK3 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr11_-_3859089 | 0.00 |
ENST00000396979.1 |
RHOG |
ras homolog family member G |
| chr1_+_186265399 | 0.00 |
ENST00000367486.3 ENST00000367484.3 ENST00000533951.1 ENST00000367482.4 ENST00000367483.4 ENST00000367485.4 ENST00000445192.2 |
PRG4 |
proteoglycan 4 |
| chr3_+_9834227 | 0.00 |
ENST00000287613.7 ENST00000397261.3 |
ARPC4 |
actin related protein 2/3 complex, subunit 4, 20kDa |
| chr12_-_6233828 | 0.00 |
ENST00000572068.1 ENST00000261405.5 |
VWF |
von Willebrand factor |
| chr5_-_79950775 | 0.00 |
ENST00000439211.2 |
DHFR |
dihydrofolate reductase |
| chr2_+_169757750 | 0.00 |
ENST00000375363.3 ENST00000429379.2 ENST00000421979.1 |
G6PC2 |
glucose-6-phosphatase, catalytic, 2 |
| chr6_+_43968306 | 0.00 |
ENST00000442114.2 ENST00000336600.5 ENST00000439969.2 |
C6orf223 |
chromosome 6 open reading frame 223 |
| chr6_-_161695074 | 0.00 |
ENST00000457520.2 ENST00000366906.5 ENST00000320285.4 |
AGPAT4 |
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr1_-_28527152 | 0.00 |
ENST00000321830.5 |
AL353354.1 |
Uncharacterized protein |
| chr16_+_14280742 | 0.00 |
ENST00000341243.5 |
MKL2 |
MKL/myocardin-like 2 |
| chr1_+_226411319 | 0.00 |
ENST00000542034.1 ENST00000366810.5 |
MIXL1 |
Mix paired-like homeobox |
| chr18_-_21977748 | 0.00 |
ENST00000399441.4 ENST00000319481.3 |
OSBPL1A |
oxysterol binding protein-like 1A |
| chr4_+_169418195 | 0.00 |
ENST00000261509.6 ENST00000335742.7 |
PALLD |
palladin, cytoskeletal associated protein |
| chr12_+_52695617 | 0.00 |
ENST00000293525.5 |
KRT86 |
keratin 86 |
| chr16_+_14280564 | 0.00 |
ENST00000572567.1 |
MKL2 |
MKL/myocardin-like 2 |
| chr12_+_25205666 | 0.00 |
ENST00000547044.1 |
LRMP |
lymphoid-restricted membrane protein |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.0 | 0.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.2 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.1 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.1 | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.3 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.2 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 | 0.1 | GO:0038188 | cholecystokinin signaling pathway(GO:0038188) |
| 0.0 | 0.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.2 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.1 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.3 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.3 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.1 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.1 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.0 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.0 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0004951 | cholecystokinin receptor activity(GO:0004951) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.2 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |