Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for VSX1
Z-value: 0.37
Transcription factors associated with VSX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
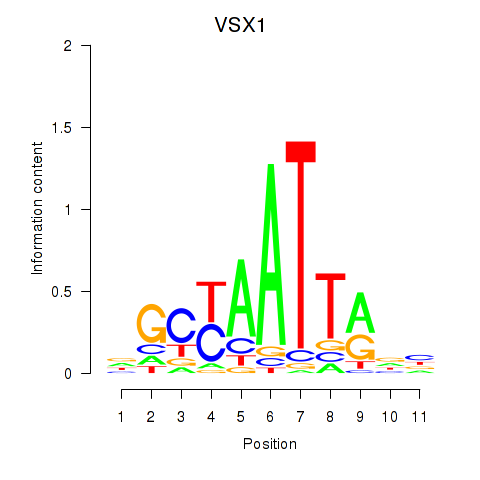
VSX1
|
ENSG00000100987.10 | VSX1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| VSX1 | hg19_v2_chr20_-_25062767_25062779 | 0.33 | 4.3e-01 | Click! |
Activity profile of VSX1 motif
Sorted Z-values of VSX1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of VSX1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_70876926 | 0.13 |
ENST00000370938.3 ENST00000346806.2 |
CTH |
cystathionase (cystathionine gamma-lyase) |
| chr1_+_70876891 | 0.12 |
ENST00000411986.2 |
CTH |
cystathionase (cystathionine gamma-lyase) |
| chr19_+_45394477 | 0.09 |
ENST00000252487.5 ENST00000405636.2 ENST00000592434.1 ENST00000426677.2 ENST00000589649.1 |
TOMM40 |
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr14_+_29236269 | 0.09 |
ENST00000313071.4 |
FOXG1 |
forkhead box G1 |
| chr6_+_31783291 | 0.08 |
ENST00000375651.5 ENST00000608703.1 ENST00000458062.2 |
HSPA1A |
heat shock 70kDa protein 1A |
| chrX_+_130192318 | 0.08 |
ENST00000370922.1 |
ARHGAP36 |
Rho GTPase activating protein 36 |
| chr5_+_98109322 | 0.07 |
ENST00000513185.1 |
RGMB |
repulsive guidance molecule family member b |
| chr1_-_197115818 | 0.07 |
ENST00000367409.4 ENST00000294732.7 |
ASPM |
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr6_-_136571400 | 0.07 |
ENST00000418509.2 ENST00000420702.1 ENST00000451457.2 |
MTFR2 |
mitochondrial fission regulator 2 |
| chrX_+_78426469 | 0.07 |
ENST00000276077.1 |
GPR174 |
G protein-coupled receptor 174 |
| chr8_+_105235572 | 0.07 |
ENST00000523362.1 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr8_-_131399110 | 0.06 |
ENST00000521426.1 |
ASAP1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
| chr9_+_12693336 | 0.06 |
ENST00000381137.2 ENST00000388918.5 |
TYRP1 |
tyrosinase-related protein 1 |
| chr3_-_112693759 | 0.06 |
ENST00000440122.2 ENST00000490004.1 |
CD200R1 |
CD200 receptor 1 |
| chr1_+_209602156 | 0.06 |
ENST00000429156.1 ENST00000366437.3 ENST00000603283.1 ENST00000431096.1 |
MIR205HG |
MIR205 host gene (non-protein coding) |
| chr5_+_53751445 | 0.06 |
ENST00000302005.1 |
HSPB3 |
heat shock 27kDa protein 3 |
| chr14_-_104181771 | 0.06 |
ENST00000554913.1 ENST00000554974.1 ENST00000553361.1 ENST00000555055.1 ENST00000555964.1 ENST00000556682.1 ENST00000445556.1 ENST00000553332.1 ENST00000352127.7 |
XRCC3 |
X-ray repair complementing defective repair in Chinese hamster cells 3 |
| chr2_+_172864490 | 0.06 |
ENST00000315796.4 |
METAP1D |
methionyl aminopeptidase type 1D (mitochondrial) |
| chr11_-_26593779 | 0.06 |
ENST00000529533.1 |
MUC15 |
mucin 15, cell surface associated |
| chr16_-_2264779 | 0.06 |
ENST00000333503.7 |
PGP |
phosphoglycolate phosphatase |
| chr6_+_31795506 | 0.06 |
ENST00000375650.3 |
HSPA1B |
heat shock 70kDa protein 1B |
| chr11_-_26593677 | 0.05 |
ENST00000527569.1 |
MUC15 |
mucin 15, cell surface associated |
| chr7_-_99699538 | 0.05 |
ENST00000343023.6 ENST00000303887.5 |
MCM7 |
minichromosome maintenance complex component 7 |
| chr5_+_33440802 | 0.05 |
ENST00000502553.1 ENST00000514259.1 ENST00000265112.3 |
TARS |
threonyl-tRNA synthetase |
| chr5_+_33441053 | 0.05 |
ENST00000541634.1 ENST00000455217.2 ENST00000414361.2 |
TARS |
threonyl-tRNA synthetase |
| chr19_+_34287751 | 0.05 |
ENST00000590771.1 ENST00000589786.1 ENST00000284006.6 ENST00000588881.1 |
KCTD15 |
potassium channel tetramerization domain containing 15 |
| chr20_+_56136136 | 0.05 |
ENST00000319441.4 ENST00000543666.1 |
PCK1 |
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr1_-_149900122 | 0.05 |
ENST00000271628.8 |
SF3B4 |
splicing factor 3b, subunit 4, 49kDa |
| chr7_+_77428066 | 0.05 |
ENST00000422959.2 ENST00000307305.8 ENST00000424760.1 |
PHTF2 |
putative homeodomain transcription factor 2 |
| chr11_+_119205222 | 0.05 |
ENST00000311413.4 |
RNF26 |
ring finger protein 26 |
| chr1_+_101003687 | 0.05 |
ENST00000315033.4 |
GPR88 |
G protein-coupled receptor 88 |
| chr7_+_77428149 | 0.05 |
ENST00000415251.2 ENST00000275575.7 |
PHTF2 |
putative homeodomain transcription factor 2 |
| chr15_+_89631647 | 0.05 |
ENST00000569550.1 ENST00000565066.1 ENST00000565973.1 |
ABHD2 |
abhydrolase domain containing 2 |
| chr3_-_195538760 | 0.05 |
ENST00000475231.1 |
MUC4 |
mucin 4, cell surface associated |
| chr19_+_50180317 | 0.05 |
ENST00000534465.1 |
PRMT1 |
protein arginine methyltransferase 1 |
| chr1_-_23520755 | 0.05 |
ENST00000314113.3 |
HTR1D |
5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
| chr21_-_34914394 | 0.04 |
ENST00000361093.5 ENST00000381815.4 |
GART |
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
| chr14_-_71276211 | 0.04 |
ENST00000381250.4 ENST00000555993.2 |
MAP3K9 |
mitogen-activated protein kinase kinase kinase 9 |
| chr3_-_195538728 | 0.04 |
ENST00000349607.4 ENST00000346145.4 |
MUC4 |
mucin 4, cell surface associated |
| chr7_+_100136811 | 0.04 |
ENST00000300176.4 ENST00000262935.4 |
AGFG2 |
ArfGAP with FG repeats 2 |
| chr19_-_13227463 | 0.04 |
ENST00000437766.1 ENST00000221504.8 |
TRMT1 |
tRNA methyltransferase 1 homolog (S. cerevisiae) |
| chr19_-_14785622 | 0.04 |
ENST00000443157.2 |
EMR3 |
egf-like module containing, mucin-like, hormone receptor-like 3 |
| chr19_-_14785674 | 0.04 |
ENST00000253673.5 |
EMR3 |
egf-like module containing, mucin-like, hormone receptor-like 3 |
| chr7_-_122339162 | 0.04 |
ENST00000340112.2 |
RNF133 |
ring finger protein 133 |
| chr4_+_87515454 | 0.04 |
ENST00000427191.2 ENST00000436978.1 ENST00000502971.1 |
PTPN13 |
protein tyrosine phosphatase, non-receptor type 13 (APO-1/CD95 (Fas)-associated phosphatase) |
| chr7_-_44613494 | 0.04 |
ENST00000431640.1 ENST00000258772.5 |
DDX56 |
DEAD (Asp-Glu-Ala-Asp) box helicase 56 |
| chr19_-_13227534 | 0.04 |
ENST00000588229.1 ENST00000357720.4 |
TRMT1 |
tRNA methyltransferase 1 homolog (S. cerevisiae) |
| chr16_+_46723552 | 0.04 |
ENST00000219097.2 ENST00000568364.2 |
ORC6 |
origin recognition complex, subunit 6 |
| chr5_+_151151471 | 0.04 |
ENST00000394123.3 ENST00000543466.1 |
G3BP1 |
GTPase activating protein (SH3 domain) binding protein 1 |
| chr16_+_640201 | 0.04 |
ENST00000563109.1 |
RAB40C |
RAB40C, member RAS oncogene family |
| chr4_+_106631966 | 0.04 |
ENST00000360505.5 ENST00000510865.1 ENST00000509336.1 |
GSTCD |
glutathione S-transferase, C-terminal domain containing |
| chr19_+_46498704 | 0.04 |
ENST00000595358.1 ENST00000594672.1 ENST00000536603.1 |
CCDC61 |
coiled-coil domain containing 61 |
| chr1_+_152943122 | 0.04 |
ENST00000328051.2 |
SPRR4 |
small proline-rich protein 4 |
| chr9_+_140135665 | 0.04 |
ENST00000340384.4 |
TUBB4B |
tubulin, beta 4B class IVb |
| chr4_+_147096837 | 0.04 |
ENST00000296581.5 ENST00000502781.1 |
LSM6 |
LSM6 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr19_-_39421377 | 0.04 |
ENST00000430193.3 ENST00000600042.1 ENST00000221431.6 |
SARS2 |
seryl-tRNA synthetase 2, mitochondrial |
| chr16_-_89883015 | 0.04 |
ENST00000563673.1 ENST00000389301.3 ENST00000568369.1 ENST00000534992.1 ENST00000389302.3 ENST00000543736.1 |
FANCA |
Fanconi anemia, complementation group A |
| chr19_+_39421556 | 0.04 |
ENST00000407800.2 ENST00000402029.3 |
MRPS12 |
mitochondrial ribosomal protein S12 |
| chr3_-_112693865 | 0.04 |
ENST00000471858.1 ENST00000295863.4 ENST00000308611.3 |
CD200R1 |
CD200 receptor 1 |
| chr15_+_90118723 | 0.04 |
ENST00000560985.1 |
TICRR |
TOPBP1-interacting checkpoint and replication regulator |
| chr1_+_206138457 | 0.04 |
ENST00000367128.3 ENST00000431655.2 |
FAM72A |
family with sequence similarity 72, member A |
| chr1_+_154947126 | 0.04 |
ENST00000368439.1 |
CKS1B |
CDC28 protein kinase regulatory subunit 1B |
| chr11_-_3818688 | 0.04 |
ENST00000355260.3 ENST00000397004.4 ENST00000397007.4 ENST00000532475.1 |
NUP98 |
nucleoporin 98kDa |
| chr1_+_154947148 | 0.04 |
ENST00000368436.1 ENST00000308987.5 |
CKS1B |
CDC28 protein kinase regulatory subunit 1B |
| chrX_-_18690210 | 0.04 |
ENST00000379984.3 |
RS1 |
retinoschisin 1 |
| chr14_-_78083112 | 0.04 |
ENST00000216484.2 |
SPTLC2 |
serine palmitoyltransferase, long chain base subunit 2 |
| chr15_+_90118685 | 0.04 |
ENST00000268138.7 |
TICRR |
TOPBP1-interacting checkpoint and replication regulator |
| chr7_+_6048856 | 0.03 |
ENST00000223029.3 ENST00000400479.2 ENST00000395236.2 |
AIMP2 |
aminoacyl tRNA synthetase complex-interacting multifunctional protein 2 |
| chr10_-_105845674 | 0.03 |
ENST00000353479.5 ENST00000369733.3 |
COL17A1 |
collagen, type XVII, alpha 1 |
| chr1_-_152386732 | 0.03 |
ENST00000271835.3 |
CRNN |
cornulin |
| chr22_+_40297079 | 0.03 |
ENST00000344138.4 ENST00000543252.1 |
GRAP2 |
GRB2-related adaptor protein 2 |
| chr11_-_3818932 | 0.03 |
ENST00000324932.7 ENST00000359171.4 |
NUP98 |
nucleoporin 98kDa |
| chr2_+_65454926 | 0.03 |
ENST00000542850.1 ENST00000377982.4 |
ACTR2 |
ARP2 actin-related protein 2 homolog (yeast) |
| chr19_-_6424783 | 0.03 |
ENST00000398148.3 |
KHSRP |
KH-type splicing regulatory protein |
| chr19_+_13001840 | 0.03 |
ENST00000222214.5 ENST00000589039.1 ENST00000591470.1 ENST00000457854.1 ENST00000422947.2 ENST00000588905.1 ENST00000587072.1 |
GCDH |
glutaryl-CoA dehydrogenase |
| chr12_-_56694142 | 0.03 |
ENST00000550655.1 ENST00000548567.1 ENST00000551430.2 ENST00000351328.3 |
CS |
citrate synthase |
| chr13_-_110438914 | 0.03 |
ENST00000375856.3 |
IRS2 |
insulin receptor substrate 2 |
| chr12_+_98909351 | 0.03 |
ENST00000343315.5 ENST00000266732.4 ENST00000393053.2 |
TMPO |
thymopoietin |
| chr2_-_201828356 | 0.03 |
ENST00000234296.2 |
ORC2 |
origin recognition complex, subunit 2 |
| chrX_+_37639264 | 0.03 |
ENST00000378588.4 |
CYBB |
cytochrome b-245, beta polypeptide |
| chr5_+_44809027 | 0.03 |
ENST00000507110.1 |
MRPS30 |
mitochondrial ribosomal protein S30 |
| chr15_-_91537723 | 0.03 |
ENST00000394249.3 ENST00000559811.1 ENST00000442656.2 ENST00000557905.1 ENST00000361919.3 |
PRC1 |
protein regulator of cytokinesis 1 |
| chr13_-_50367057 | 0.03 |
ENST00000261667.3 |
KPNA3 |
karyopherin alpha 3 (importin alpha 4) |
| chr6_+_33422343 | 0.03 |
ENST00000395064.2 |
ZBTB9 |
zinc finger and BTB domain containing 9 |
| chr17_+_79650962 | 0.03 |
ENST00000329138.4 |
HGS |
hepatocyte growth factor-regulated tyrosine kinase substrate |
| chr12_-_6960407 | 0.03 |
ENST00000540683.1 ENST00000229265.6 ENST00000535406.1 ENST00000422785.3 |
CDCA3 |
cell division cycle associated 3 |
| chr15_-_43559055 | 0.03 |
ENST00000220420.5 ENST00000349114.4 |
TGM5 |
transglutaminase 5 |
| chr12_+_106751436 | 0.03 |
ENST00000228347.4 |
POLR3B |
polymerase (RNA) III (DNA directed) polypeptide B |
| chr12_+_98909260 | 0.03 |
ENST00000556029.1 |
TMPO |
thymopoietin |
| chr12_-_6961050 | 0.03 |
ENST00000538862.2 |
CDCA3 |
cell division cycle associated 3 |
| chr5_+_159343688 | 0.03 |
ENST00000306675.3 |
ADRA1B |
adrenoceptor alpha 1B |
| chrX_+_151999511 | 0.03 |
ENST00000370274.3 ENST00000440023.1 ENST00000432467.1 |
NSDHL |
NAD(P) dependent steroid dehydrogenase-like |
| chr5_-_82969405 | 0.03 |
ENST00000510978.1 |
HAPLN1 |
hyaluronan and proteoglycan link protein 1 |
| chrM_+_4431 | 0.03 |
ENST00000361453.3 |
MT-ND2 |
mitochondrially encoded NADH dehydrogenase 2 |
| chrM_+_10758 | 0.03 |
ENST00000361381.2 |
MT-ND4 |
mitochondrially encoded NADH dehydrogenase 4 |
| chr11_-_121986923 | 0.03 |
ENST00000560104.1 |
BLID |
BH3-like motif containing, cell death inducer |
| chr3_+_186288454 | 0.03 |
ENST00000265028.3 |
DNAJB11 |
DnaJ (Hsp40) homolog, subfamily B, member 11 |
| chr4_+_40198527 | 0.03 |
ENST00000381799.5 |
RHOH |
ras homolog family member H |
| chr12_+_81110684 | 0.03 |
ENST00000228644.3 |
MYF5 |
myogenic factor 5 |
| chr6_-_26032288 | 0.03 |
ENST00000244661.2 |
HIST1H3B |
histone cluster 1, H3b |
| chr14_+_58711539 | 0.03 |
ENST00000216455.4 ENST00000412908.2 ENST00000557508.1 |
PSMA3 |
proteasome (prosome, macropain) subunit, alpha type, 3 |
| chr7_-_148725733 | 0.03 |
ENST00000286091.4 |
PDIA4 |
protein disulfide isomerase family A, member 4 |
| chr12_-_49075941 | 0.03 |
ENST00000553086.1 ENST00000548304.1 |
KANSL2 |
KAT8 regulatory NSL complex subunit 2 |
| chr2_-_192711968 | 0.03 |
ENST00000304141.4 |
SDPR |
serum deprivation response |
| chr2_-_24270217 | 0.03 |
ENST00000295148.4 ENST00000406895.3 |
C2orf44 |
chromosome 2 open reading frame 44 |
| chrX_-_133931164 | 0.03 |
ENST00000370790.1 ENST00000298090.6 |
FAM122B |
family with sequence similarity 122B |
| chr2_+_153574428 | 0.03 |
ENST00000326446.5 |
ARL6IP6 |
ADP-ribosylation-like factor 6 interacting protein 6 |
| chr1_-_78444776 | 0.03 |
ENST00000370767.1 ENST00000421641.1 |
FUBP1 |
far upstream element (FUSE) binding protein 1 |
| chr11_+_57480046 | 0.03 |
ENST00000378312.4 ENST00000278422.4 |
TMX2 |
thioredoxin-related transmembrane protein 2 |
| chr19_+_46195895 | 0.03 |
ENST00000366382.4 |
QPCTL |
glutaminyl-peptide cyclotransferase-like |
| chr1_-_35658736 | 0.03 |
ENST00000357214.5 |
SFPQ |
splicing factor proline/glutamine-rich |
| chr11_+_12308447 | 0.03 |
ENST00000256186.2 |
MICALCL |
MICAL C-terminal like |
| chr2_-_227050079 | 0.03 |
ENST00000423838.1 |
AC068138.1 |
AC068138.1 |
| chr6_-_27860956 | 0.03 |
ENST00000359611.2 |
HIST1H2AM |
histone cluster 1, H2am |
| chr5_+_179125368 | 0.03 |
ENST00000502296.1 ENST00000504734.1 ENST00000415618.2 |
CANX |
calnexin |
| chr5_+_151151504 | 0.03 |
ENST00000356245.3 ENST00000507878.2 |
G3BP1 |
GTPase activating protein (SH3 domain) binding protein 1 |
| chr3_-_148939835 | 0.03 |
ENST00000264613.6 |
CP |
ceruloplasmin (ferroxidase) |
| chr19_-_54663473 | 0.03 |
ENST00000222224.3 |
LENG1 |
leukocyte receptor cluster (LRC) member 1 |
| chr17_-_40829026 | 0.03 |
ENST00000412503.1 |
PLEKHH3 |
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr17_-_46799872 | 0.03 |
ENST00000290294.3 |
PRAC1 |
prostate cancer susceptibility candidate 1 |
| chr12_+_110011571 | 0.03 |
ENST00000539696.1 ENST00000228510.3 ENST00000392727.3 |
MVK |
mevalonate kinase |
| chrX_+_37639302 | 0.03 |
ENST00000545017.1 ENST00000536160.1 |
CYBB |
cytochrome b-245, beta polypeptide |
| chr1_-_78444738 | 0.02 |
ENST00000436586.2 ENST00000370768.2 |
FUBP1 |
far upstream element (FUSE) binding protein 1 |
| chrX_+_70503433 | 0.02 |
ENST00000276079.8 ENST00000373856.3 ENST00000373841.1 ENST00000420903.1 |
NONO |
non-POU domain containing, octamer-binding |
| chr3_-_138048682 | 0.02 |
ENST00000383180.2 |
NME9 |
NME/NM23 family member 9 |
| chr10_+_74451883 | 0.02 |
ENST00000373053.3 ENST00000357157.6 |
MCU |
mitochondrial calcium uniporter |
| chr19_+_10828795 | 0.02 |
ENST00000389253.4 ENST00000355667.6 ENST00000408974.4 |
DNM2 |
dynamin 2 |
| chr2_+_131100423 | 0.02 |
ENST00000409935.1 ENST00000409649.1 ENST00000428740.1 |
IMP4 |
IMP4, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
| chr6_+_111408698 | 0.02 |
ENST00000368851.5 |
SLC16A10 |
solute carrier family 16 (aromatic amino acid transporter), member 10 |
| chr11_+_125774258 | 0.02 |
ENST00000263576.6 |
DDX25 |
DEAD (Asp-Glu-Ala-Asp) box helicase 25 |
| chr19_+_19496728 | 0.02 |
ENST00000537887.1 ENST00000417582.2 |
GATAD2A |
GATA zinc finger domain containing 2A |
| chrX_+_70503037 | 0.02 |
ENST00000535149.1 |
NONO |
non-POU domain containing, octamer-binding |
| chr5_+_179125907 | 0.02 |
ENST00000247461.4 ENST00000452673.2 ENST00000502498.1 ENST00000507307.1 ENST00000513246.1 ENST00000502673.1 ENST00000506654.1 ENST00000512607.2 ENST00000510810.1 |
CANX |
calnexin |
| chr1_+_42846443 | 0.02 |
ENST00000410070.2 ENST00000431473.3 |
RIMKLA |
ribosomal modification protein rimK-like family member A |
| chr11_+_125774362 | 0.02 |
ENST00000530414.1 ENST00000530129.2 |
DDX25 |
DEAD (Asp-Glu-Ala-Asp) box helicase 25 |
| chr17_-_5323480 | 0.02 |
ENST00000573584.1 |
NUP88 |
nucleoporin 88kDa |
| chr19_-_2256405 | 0.02 |
ENST00000300961.6 |
JSRP1 |
junctional sarcoplasmic reticulum protein 1 |
| chr6_+_130339710 | 0.02 |
ENST00000526087.1 ENST00000533560.1 ENST00000361794.2 |
L3MBTL3 |
l(3)mbt-like 3 (Drosophila) |
| chr4_+_166248775 | 0.02 |
ENST00000261507.6 ENST00000507013.1 ENST00000393766.2 ENST00000504317.1 |
MSMO1 |
methylsterol monooxygenase 1 |
| chr17_-_5322786 | 0.02 |
ENST00000225696.4 |
NUP88 |
nucleoporin 88kDa |
| chr19_-_15236470 | 0.02 |
ENST00000533747.1 ENST00000598709.1 ENST00000534378.1 |
ILVBL |
ilvB (bacterial acetolactate synthase)-like |
| chrX_+_19373700 | 0.02 |
ENST00000379804.1 |
PDHA1 |
pyruvate dehydrogenase (lipoamide) alpha 1 |
| chr6_+_31707725 | 0.02 |
ENST00000375755.3 ENST00000375742.3 ENST00000375750.3 ENST00000425703.1 ENST00000534153.4 ENST00000375703.3 ENST00000375740.3 |
MSH5 |
mutS homolog 5 |
| chr11_-_95522639 | 0.02 |
ENST00000536839.1 |
FAM76B |
family with sequence similarity 76, member B |
| chr6_+_25754927 | 0.02 |
ENST00000377905.4 ENST00000439485.2 |
SLC17A4 |
solute carrier family 17, member 4 |
| chr3_-_165555200 | 0.02 |
ENST00000479451.1 ENST00000540653.1 ENST00000488954.1 ENST00000264381.3 |
BCHE |
butyrylcholinesterase |
| chr3_+_130279178 | 0.02 |
ENST00000358511.6 ENST00000453409.2 |
COL6A6 |
collagen, type VI, alpha 6 |
| chr2_-_153574480 | 0.02 |
ENST00000410080.1 |
PRPF40A |
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr19_-_11266471 | 0.02 |
ENST00000592540.1 |
SPC24 |
SPC24, NDC80 kinetochore complex component |
| chr4_-_87028478 | 0.02 |
ENST00000515400.1 ENST00000395157.3 |
MAPK10 |
mitogen-activated protein kinase 10 |
| chr18_-_33709268 | 0.02 |
ENST00000269187.5 ENST00000590986.1 ENST00000440549.2 |
SLC39A6 |
solute carrier family 39 (zinc transporter), member 6 |
| chr19_+_10828724 | 0.02 |
ENST00000585892.1 ENST00000314646.5 ENST00000359692.6 |
DNM2 |
dynamin 2 |
| chr8_+_120428546 | 0.02 |
ENST00000259526.3 |
NOV |
nephroblastoma overexpressed |
| chr13_-_19755975 | 0.02 |
ENST00000400113.3 |
TUBA3C |
tubulin, alpha 3c |
| chr7_-_77427676 | 0.02 |
ENST00000257663.3 |
TMEM60 |
transmembrane protein 60 |
| chr19_+_50180409 | 0.02 |
ENST00000391851.4 |
PRMT1 |
protein arginine methyltransferase 1 |
| chr19_+_11485333 | 0.02 |
ENST00000312423.2 |
SWSAP1 |
SWIM-type zinc finger 7 associated protein 1 |
| chr11_-_33913708 | 0.02 |
ENST00000257818.2 |
LMO2 |
LIM domain only 2 (rhombotin-like 1) |
| chr14_-_73925225 | 0.02 |
ENST00000356296.4 ENST00000355058.3 ENST00000359560.3 ENST00000557597.1 ENST00000554394.1 ENST00000555238.1 ENST00000535282.1 ENST00000555987.1 ENST00000555394.1 ENST00000554546.1 |
NUMB |
numb homolog (Drosophila) |
| chr3_+_100120441 | 0.02 |
ENST00000489752.1 |
LNP1 |
leukemia NUP98 fusion partner 1 |
| chr3_-_42003479 | 0.02 |
ENST00000420927.1 |
ULK4 |
unc-51 like kinase 4 |
| chr19_+_54619125 | 0.02 |
ENST00000445811.1 ENST00000419967.1 ENST00000445124.1 ENST00000447810.1 |
PRPF31 |
pre-mRNA processing factor 31 |
| chr7_+_55433131 | 0.02 |
ENST00000254770.2 |
LANCL2 |
LanC lantibiotic synthetase component C-like 2 (bacterial) |
| chr5_-_13944652 | 0.02 |
ENST00000265104.4 |
DNAH5 |
dynein, axonemal, heavy chain 5 |
| chr19_+_54641444 | 0.02 |
ENST00000221232.5 ENST00000358389.3 |
CNOT3 |
CCR4-NOT transcription complex, subunit 3 |
| chr1_-_204380919 | 0.02 |
ENST00000367188.4 |
PPP1R15B |
protein phosphatase 1, regulatory subunit 15B |
| chr17_-_1733114 | 0.02 |
ENST00000305513.7 |
SMYD4 |
SET and MYND domain containing 4 |
| chr1_-_234667504 | 0.02 |
ENST00000421207.1 ENST00000435574.1 |
RP5-855F14.1 |
RP5-855F14.1 |
| chr19_+_4007644 | 0.02 |
ENST00000262971.2 |
PIAS4 |
protein inhibitor of activated STAT, 4 |
| chr1_-_6052463 | 0.02 |
ENST00000378156.4 |
NPHP4 |
nephronophthisis 4 |
| chr4_-_39979576 | 0.02 |
ENST00000303538.8 ENST00000503396.1 |
PDS5A |
PDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae) |
| chr19_+_19496624 | 0.02 |
ENST00000494516.2 ENST00000360315.3 ENST00000252577.5 |
GATAD2A |
GATA zinc finger domain containing 2A |
| chr2_-_55276320 | 0.02 |
ENST00000357376.3 |
RTN4 |
reticulon 4 |
| chr17_+_45726803 | 0.02 |
ENST00000535458.2 ENST00000583648.1 |
KPNB1 |
karyopherin (importin) beta 1 |
| chr11_-_122933043 | 0.02 |
ENST00000534624.1 ENST00000453788.2 ENST00000527387.1 |
HSPA8 |
heat shock 70kDa protein 8 |
| chr18_-_33702078 | 0.02 |
ENST00000586829.1 |
SLC39A6 |
solute carrier family 39 (zinc transporter), member 6 |
| chr17_-_47785504 | 0.02 |
ENST00000514907.1 ENST00000503334.1 ENST00000508520.1 |
SLC35B1 |
solute carrier family 35, member B1 |
| chr11_+_128563652 | 0.02 |
ENST00000527786.2 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr3_+_179280668 | 0.02 |
ENST00000429709.2 ENST00000450518.2 ENST00000392662.1 ENST00000490364.1 |
ACTL6A |
actin-like 6A |
| chr6_+_36164487 | 0.02 |
ENST00000357641.6 |
BRPF3 |
bromodomain and PHD finger containing, 3 |
| chr19_-_47137942 | 0.02 |
ENST00000300873.4 |
GNG8 |
guanine nucleotide binding protein (G protein), gamma 8 |
| chr19_-_7694417 | 0.02 |
ENST00000358368.4 ENST00000534844.1 |
XAB2 |
XPA binding protein 2 |
| chr5_-_49737184 | 0.02 |
ENST00000508934.1 ENST00000303221.5 |
EMB |
embigin |
| chr3_-_119813264 | 0.02 |
ENST00000264235.8 |
GSK3B |
glycogen synthase kinase 3 beta |
| chr11_+_43702322 | 0.02 |
ENST00000395700.4 |
HSD17B12 |
hydroxysteroid (17-beta) dehydrogenase 12 |
| chr20_+_37554955 | 0.02 |
ENST00000217429.4 |
FAM83D |
family with sequence similarity 83, member D |
| chr20_+_16729003 | 0.02 |
ENST00000246081.2 |
OTOR |
otoraplin |
| chr1_+_40713573 | 0.02 |
ENST00000372766.3 |
TMCO2 |
transmembrane and coiled-coil domains 2 |
| chr20_+_44441304 | 0.02 |
ENST00000352551.5 |
UBE2C |
ubiquitin-conjugating enzyme E2C |
| chr17_-_74733404 | 0.02 |
ENST00000508921.3 ENST00000583836.1 ENST00000358156.6 ENST00000392485.2 ENST00000359995.5 |
SRSF2 |
serine/arginine-rich splicing factor 2 |
| chr7_+_138145145 | 0.02 |
ENST00000415680.2 |
TRIM24 |
tripartite motif containing 24 |
| chr9_+_1050331 | 0.02 |
ENST00000382255.3 ENST00000382251.3 ENST00000412350.2 |
DMRT2 |
doublesex and mab-3 related transcription factor 2 |
| chrX_-_24665353 | 0.02 |
ENST00000379144.2 |
PCYT1B |
phosphate cytidylyltransferase 1, choline, beta |
| chr19_+_1205740 | 0.02 |
ENST00000326873.7 |
STK11 |
serine/threonine kinase 11 |
| chr19_-_15236173 | 0.02 |
ENST00000527093.1 |
ILVBL |
ilvB (bacterial acetolactate synthase)-like |
| chr20_+_44441215 | 0.02 |
ENST00000356455.4 ENST00000405520.1 |
UBE2C |
ubiquitin-conjugating enzyme E2C |
| chr3_+_47324424 | 0.02 |
ENST00000437353.1 ENST00000232766.5 ENST00000455924.2 |
KLHL18 |
kelch-like family member 18 |
| chr17_-_45266542 | 0.02 |
ENST00000531206.1 ENST00000527547.1 ENST00000446365.2 ENST00000575483.1 ENST00000066544.3 |
CDC27 |
cell division cycle 27 |
| chr11_+_63754294 | 0.02 |
ENST00000543988.1 |
OTUB1 |
OTU domain, ubiquitin aldehyde binding 1 |
| chr2_-_101925055 | 0.02 |
ENST00000295317.3 |
RNF149 |
ring finger protein 149 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.0 | 0.1 | GO:0070426 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 0.0 | 0.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 0.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.1 | GO:0071139 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.0 | 0.0 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.1 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.0 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.0 | GO:1904000 | positive regulation of eating behavior(GO:1904000) |
| 0.0 | 0.0 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.0 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.0 | 0.0 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.1 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 0.1 | GO:1903525 | regulation of membrane tubulation(GO:1903525) positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.0 | GO:0015785 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.0 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.0 | 0.0 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.1 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.2 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.1 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.0 | 0.0 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.1 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.1 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.0 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.0 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |