Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
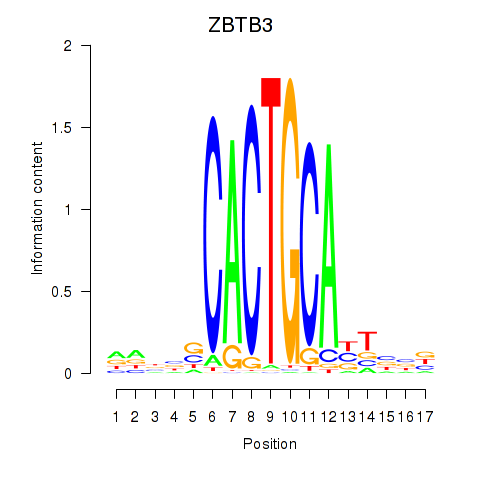
Results for ZBTB3
Z-value: 0.50
Transcription factors associated with ZBTB3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB3
|
ENSG00000185670.7 | ZBTB3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB3 | hg19_v2_chr11_-_62521614_62521660 | 0.54 | 1.7e-01 | Click! |
Activity profile of ZBTB3 motif
Sorted Z-values of ZBTB3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_92414055 | 1.08 |
ENST00000342058.4 |
FBLN5 |
fibulin 5 |
| chr1_+_159141397 | 1.03 |
ENST00000368124.4 ENST00000368125.4 ENST00000416746.1 |
CADM3 |
cell adhesion molecule 3 |
| chr14_-_92413353 | 1.03 |
ENST00000556154.1 |
FBLN5 |
fibulin 5 |
| chr14_-_92413727 | 0.89 |
ENST00000267620.10 |
FBLN5 |
fibulin 5 |
| chr2_+_202899310 | 0.46 |
ENST00000286201.1 |
FZD7 |
frizzled family receptor 7 |
| chr11_-_82782861 | 0.31 |
ENST00000524635.1 ENST00000526205.1 ENST00000527633.1 ENST00000533486.1 ENST00000533276.2 |
RAB30 |
RAB30, member RAS oncogene family |
| chr11_-_82782952 | 0.26 |
ENST00000534141.1 |
RAB30 |
RAB30, member RAS oncogene family |
| chr1_+_211433275 | 0.21 |
ENST00000367005.4 |
RCOR3 |
REST corepressor 3 |
| chr4_+_166300084 | 0.21 |
ENST00000402744.4 |
CPE |
carboxypeptidase E |
| chr1_+_110036699 | 0.21 |
ENST00000496961.1 ENST00000533024.1 ENST00000310611.4 ENST00000527072.1 ENST00000420578.2 ENST00000528785.1 |
CYB561D1 |
cytochrome b561 family, member D1 |
| chr1_+_110036728 | 0.20 |
ENST00000369868.3 ENST00000430195.2 |
CYB561D1 |
cytochrome b561 family, member D1 |
| chrX_-_118827333 | 0.19 |
ENST00000360156.7 ENST00000354228.4 ENST00000489216.1 ENST00000354416.3 ENST00000394610.1 ENST00000343984.5 |
SEPT6 |
septin 6 |
| chr5_+_170288856 | 0.18 |
ENST00000523189.1 |
RANBP17 |
RAN binding protein 17 |
| chr19_-_40023450 | 0.17 |
ENST00000326282.4 |
EID2B |
EP300 interacting inhibitor of differentiation 2B |
| chr2_-_40679186 | 0.16 |
ENST00000406785.2 |
SLC8A1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr8_+_22424551 | 0.16 |
ENST00000523348.1 |
SORBS3 |
sorbin and SH3 domain containing 3 |
| chr4_-_5894777 | 0.16 |
ENST00000324989.7 |
CRMP1 |
collapsin response mediator protein 1 |
| chr5_+_112074029 | 0.16 |
ENST00000512211.2 |
APC |
adenomatous polyposis coli |
| chr7_-_38403077 | 0.15 |
ENST00000426402.2 |
TRGV2 |
T cell receptor gamma variable 2 |
| chr19_-_6670128 | 0.15 |
ENST00000245912.3 |
TNFSF14 |
tumor necrosis factor (ligand) superfamily, member 14 |
| chr7_-_38394118 | 0.13 |
ENST00000390345.2 |
TRGV4 |
T cell receptor gamma variable 4 |
| chrX_+_134166333 | 0.13 |
ENST00000257013.7 |
FAM127A |
family with sequence similarity 127, member A |
| chr11_-_130786333 | 0.13 |
ENST00000533214.1 ENST00000528555.1 ENST00000530356.1 ENST00000539184.1 |
SNX19 |
sorting nexin 19 |
| chr4_-_17783135 | 0.12 |
ENST00000265018.3 |
FAM184B |
family with sequence similarity 184, member B |
| chr16_+_53164833 | 0.12 |
ENST00000564845.1 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
| chr8_-_42623747 | 0.11 |
ENST00000534622.1 |
CHRNA6 |
cholinergic receptor, nicotinic, alpha 6 (neuronal) |
| chr8_-_42623924 | 0.11 |
ENST00000276410.2 |
CHRNA6 |
cholinergic receptor, nicotinic, alpha 6 (neuronal) |
| chr17_-_39623681 | 0.11 |
ENST00000225899.3 |
KRT32 |
keratin 32 |
| chr7_+_134576317 | 0.10 |
ENST00000424922.1 ENST00000495522.1 |
CALD1 |
caldesmon 1 |
| chr5_+_140514782 | 0.10 |
ENST00000231134.5 |
PCDHB5 |
protocadherin beta 5 |
| chr2_+_241631262 | 0.10 |
ENST00000337801.4 ENST00000429564.1 |
AQP12A |
aquaporin 12A |
| chr1_-_236046872 | 0.09 |
ENST00000536965.1 |
LYST |
lysosomal trafficking regulator |
| chr14_-_90085458 | 0.09 |
ENST00000345097.4 ENST00000555855.1 ENST00000555353.1 |
FOXN3 |
forkhead box N3 |
| chr6_+_44126545 | 0.09 |
ENST00000532171.1 ENST00000398776.1 ENST00000542245.1 |
CAPN11 |
calpain 11 |
| chr12_+_48577366 | 0.09 |
ENST00000316554.3 |
C12orf68 |
chromosome 12 open reading frame 68 |
| chr17_+_75123947 | 0.09 |
ENST00000586429.1 |
SEC14L1 |
SEC14-like 1 (S. cerevisiae) |
| chr1_-_169555779 | 0.08 |
ENST00000367797.3 ENST00000367796.3 |
F5 |
coagulation factor V (proaccelerin, labile factor) |
| chr1_+_155146318 | 0.08 |
ENST00000368385.4 ENST00000545012.1 ENST00000392451.2 ENST00000368383.3 ENST00000368382.1 ENST00000334634.4 |
TRIM46 |
tripartite motif containing 46 |
| chr15_-_74659978 | 0.07 |
ENST00000541301.1 ENST00000416978.1 ENST00000268053.6 |
CYP11A1 |
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chrX_+_152224766 | 0.07 |
ENST00000370265.4 ENST00000447306.1 |
PNMA3 |
paraneoplastic Ma antigen 3 |
| chr19_-_19006920 | 0.07 |
ENST00000429504.2 ENST00000427170.2 |
CERS1 |
ceramide synthase 1 |
| chr18_+_32073253 | 0.06 |
ENST00000283365.9 ENST00000596745.1 ENST00000315456.6 |
DTNA |
dystrobrevin, alpha |
| chr2_+_42396574 | 0.06 |
ENST00000401738.3 |
EML4 |
echinoderm microtubule associated protein like 4 |
| chr5_-_115177247 | 0.06 |
ENST00000500945.2 |
ATG12 |
autophagy related 12 |
| chr6_-_41122063 | 0.06 |
ENST00000426005.2 ENST00000437044.2 ENST00000373127.4 |
TREML1 |
triggering receptor expressed on myeloid cells-like 1 |
| chrX_+_118708493 | 0.06 |
ENST00000371558.2 |
UBE2A |
ubiquitin-conjugating enzyme E2A |
| chr2_-_154335300 | 0.05 |
ENST00000325926.3 |
RPRM |
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr7_+_134576151 | 0.05 |
ENST00000393118.2 |
CALD1 |
caldesmon 1 |
| chr12_+_123949053 | 0.05 |
ENST00000350887.5 |
SNRNP35 |
small nuclear ribonucleoprotein 35kDa (U11/U12) |
| chr11_-_130786400 | 0.05 |
ENST00000265909.4 |
SNX19 |
sorting nexin 19 |
| chr2_-_40679148 | 0.05 |
ENST00000417271.1 |
SLC8A1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr3_-_195163803 | 0.05 |
ENST00000326793.6 |
ACAP2 |
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
| chr5_-_94417339 | 0.04 |
ENST00000429576.2 ENST00000508509.1 ENST00000510732.1 |
MCTP1 |
multiple C2 domains, transmembrane 1 |
| chr3_-_149688502 | 0.04 |
ENST00000481767.1 ENST00000475518.1 |
PFN2 |
profilin 2 |
| chr19_+_47852538 | 0.04 |
ENST00000328771.4 |
DHX34 |
DEAH (Asp-Glu-Ala-His) box polypeptide 34 |
| chr1_-_149908710 | 0.04 |
ENST00000439741.2 ENST00000361405.6 ENST00000406732.3 |
MTMR11 |
myotubularin related protein 11 |
| chr3_-_149688655 | 0.04 |
ENST00000461930.1 ENST00000423691.2 ENST00000490975.1 ENST00000461868.1 ENST00000452853.2 |
PFN2 |
profilin 2 |
| chrX_+_152240819 | 0.04 |
ENST00000421798.3 ENST00000535416.1 |
PNMA6C PNMA6A |
paraneoplastic Ma antigen family member 6C paraneoplastic Ma antigen family member 6A |
| chr16_+_89787393 | 0.04 |
ENST00000289816.5 ENST00000568064.1 |
ZNF276 |
zinc finger protein 276 |
| chr10_-_118032979 | 0.04 |
ENST00000355422.6 |
GFRA1 |
GDNF family receptor alpha 1 |
| chr15_+_64428529 | 0.04 |
ENST00000560861.1 |
SNX1 |
sorting nexin 1 |
| chr20_+_62289640 | 0.04 |
ENST00000508582.2 ENST00000360203.5 ENST00000356810.4 |
RTEL1 |
regulator of telomere elongation helicase 1 |
| chr2_+_42396472 | 0.04 |
ENST00000318522.5 ENST00000402711.2 |
EML4 |
echinoderm microtubule associated protein like 4 |
| chr16_-_19896220 | 0.04 |
ENST00000562469.1 ENST00000300571.2 |
GPRC5B |
G protein-coupled receptor, family C, group 5, member B |
| chr19_-_55549624 | 0.03 |
ENST00000417454.1 ENST00000310373.3 ENST00000333884.2 |
GP6 |
glycoprotein VI (platelet) |
| chr6_+_123100620 | 0.03 |
ENST00000368444.3 |
FABP7 |
fatty acid binding protein 7, brain |
| chr11_+_67007518 | 0.03 |
ENST00000530342.1 ENST00000308783.5 |
KDM2A |
lysine (K)-specific demethylase 2A |
| chr12_+_54718904 | 0.03 |
ENST00000262061.2 ENST00000549043.1 ENST00000552218.1 ENST00000553231.1 ENST00000552362.1 ENST00000455864.2 ENST00000416254.2 ENST00000549116.1 ENST00000551779.1 |
COPZ1 |
coatomer protein complex, subunit zeta 1 |
| chrX_+_152338301 | 0.03 |
ENST00000453825.2 |
PNMA6A |
paraneoplastic Ma antigen family member 6A |
| chr1_+_160160346 | 0.03 |
ENST00000368078.3 |
CASQ1 |
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr2_-_136594740 | 0.03 |
ENST00000264162.2 |
LCT |
lactase |
| chr16_+_87636474 | 0.03 |
ENST00000284262.2 |
JPH3 |
junctophilin 3 |
| chrX_+_118708517 | 0.03 |
ENST00000346330.3 |
UBE2A |
ubiquitin-conjugating enzyme E2A |
| chr2_-_25565377 | 0.03 |
ENST00000264709.3 ENST00000406659.3 |
DNMT3A |
DNA (cytosine-5-)-methyltransferase 3 alpha |
| chr3_-_53878644 | 0.03 |
ENST00000481668.1 ENST00000467802.1 |
CHDH |
choline dehydrogenase |
| chr5_+_138609441 | 0.03 |
ENST00000509990.1 ENST00000506147.1 ENST00000512107.1 |
MATR3 |
matrin 3 |
| chr7_+_62809239 | 0.03 |
ENST00000456890.1 |
AC006455.1 |
AC006455.1 |
| chr19_-_19006890 | 0.03 |
ENST00000247005.6 |
GDF1 |
growth differentiation factor 1 |
| chr12_-_62586543 | 0.03 |
ENST00000416284.3 |
FAM19A2 |
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2 |
| chr21_+_45209394 | 0.02 |
ENST00000497547.1 |
RRP1 |
ribosomal RNA processing 1 |
| chr5_+_75379224 | 0.02 |
ENST00000322285.7 |
SV2C |
synaptic vesicle glycoprotein 2C |
| chr17_-_7531121 | 0.02 |
ENST00000573566.1 ENST00000269298.5 |
SAT2 |
spermidine/spermine N1-acetyltransferase family member 2 |
| chr6_+_12958137 | 0.02 |
ENST00000457702.2 ENST00000379345.2 |
PHACTR1 |
phosphatase and actin regulator 1 |
| chr7_+_100081542 | 0.02 |
ENST00000300179.2 ENST00000423930.1 |
NYAP1 |
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 1 |
| chr19_+_16187085 | 0.02 |
ENST00000300933.4 |
TPM4 |
tropomyosin 4 |
| chr3_-_149688896 | 0.02 |
ENST00000239940.7 |
PFN2 |
profilin 2 |
| chr11_+_64692143 | 0.02 |
ENST00000164133.2 ENST00000532850.1 |
PPP2R5B |
protein phosphatase 2, regulatory subunit B', beta |
| chr9_-_34637718 | 0.02 |
ENST00000378892.1 ENST00000277010.4 |
SIGMAR1 |
sigma non-opioid intracellular receptor 1 |
| chr5_+_115177178 | 0.02 |
ENST00000316788.7 |
AP3S1 |
adaptor-related protein complex 3, sigma 1 subunit |
| chr11_-_18548426 | 0.02 |
ENST00000357193.3 ENST00000536719.1 |
TSG101 |
tumor susceptibility 101 |
| chr16_+_28986085 | 0.01 |
ENST00000565975.1 ENST00000311008.11 ENST00000323081.8 ENST00000334536.8 |
SPNS1 |
spinster homolog 1 (Drosophila) |
| chr11_+_82783097 | 0.01 |
ENST00000501011.2 ENST00000527627.1 ENST00000526795.1 ENST00000533528.1 ENST00000533708.1 ENST00000534499.1 |
RAB30-AS1 |
RAB30 antisense RNA 1 (head to head) |
| chr11_-_67980744 | 0.01 |
ENST00000401547.2 ENST00000453170.1 ENST00000304363.4 |
SUV420H1 |
suppressor of variegation 4-20 homolog 1 (Drosophila) |
| chr12_+_8185288 | 0.01 |
ENST00000162391.3 |
FOXJ2 |
forkhead box J2 |
| chr7_+_23719749 | 0.01 |
ENST00000409192.3 ENST00000344962.4 ENST00000409653.1 ENST00000409994.3 |
FAM221A |
family with sequence similarity 221, member A |
| chr6_-_46138676 | 0.01 |
ENST00000371383.2 ENST00000230565.3 |
ENPP5 |
ectonucleotide pyrophosphatase/phosphodiesterase 5 (putative) |
| chr5_+_76506706 | 0.01 |
ENST00000340978.3 ENST00000346042.3 ENST00000264917.5 ENST00000342343.4 ENST00000333194.4 |
PDE8B |
phosphodiesterase 8B |
| chr3_+_178276488 | 0.01 |
ENST00000432997.1 ENST00000455865.1 |
KCNMB2 |
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chrX_+_11776278 | 0.01 |
ENST00000312196.4 ENST00000337339.2 |
MSL3 |
male-specific lethal 3 homolog (Drosophila) |
| chr9_+_74526532 | 0.01 |
ENST00000486911.2 |
C9orf85 |
chromosome 9 open reading frame 85 |
| chr20_+_54987305 | 0.01 |
ENST00000371336.3 ENST00000434344.1 |
CASS4 |
Cas scaffolding protein family member 4 |
| chr9_+_125132803 | 0.01 |
ENST00000540753.1 |
PTGS1 |
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr16_-_2185899 | 0.01 |
ENST00000262304.4 ENST00000423118.1 |
PKD1 |
polycystic kidney disease 1 (autosomal dominant) |
| chr1_-_154458520 | 0.01 |
ENST00000486773.1 |
SHE |
Src homology 2 domain containing E |
| chr11_+_73661364 | 0.01 |
ENST00000339764.1 |
DNAJB13 |
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr16_+_28986134 | 0.01 |
ENST00000352260.7 |
SPNS1 |
spinster homolog 1 (Drosophila) |
| chr14_-_106725723 | 0.00 |
ENST00000390609.2 |
IGHV3-23 |
immunoglobulin heavy variable 3-23 |
| chr1_-_155270770 | 0.00 |
ENST00000392414.3 |
PKLR |
pyruvate kinase, liver and RBC |
| chr19_-_39926268 | 0.00 |
ENST00000599705.1 |
RPS16 |
ribosomal protein S16 |
| chr3_-_49131788 | 0.00 |
ENST00000395443.2 ENST00000411682.1 |
QRICH1 |
glutamine-rich 1 |
| chr5_+_74807581 | 0.00 |
ENST00000241436.4 ENST00000352007.5 |
POLK |
polymerase (DNA directed) kappa |
| chr1_+_154229547 | 0.00 |
ENST00000428595.1 |
UBAP2L |
ubiquitin associated protein 2-like |
| chr1_+_28285973 | 0.00 |
ENST00000373884.5 |
XKR8 |
XK, Kell blood group complex subunit-related family, member 8 |
| chr5_+_74807886 | 0.00 |
ENST00000514296.1 |
POLK |
polymerase (DNA directed) kappa |
| chr6_+_29274403 | 0.00 |
ENST00000377160.2 |
OR14J1 |
olfactory receptor, family 14, subfamily J, member 1 |
| chr20_+_54987168 | 0.00 |
ENST00000360314.3 |
CASS4 |
Cas scaffolding protein family member 4 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.0 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.5 | GO:0009996 | negative regulation of cell fate specification(GO:0009996) |
| 0.1 | 0.2 | GO:1904530 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.1 | 0.2 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.2 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 0.1 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0072720 | cellular response to mycotoxin(GO:0036146) response to dithiothreitol(GO:0072720) |
| 0.0 | 0.2 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.2 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.1 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.0 | 1.0 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.0 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.0 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 0.2 | GO:0005940 | septin ring(GO:0005940) septin collar(GO:0032173) |
| 0.0 | 0.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.1 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |
| 0.0 | 0.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 3.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.1 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 3.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |