Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
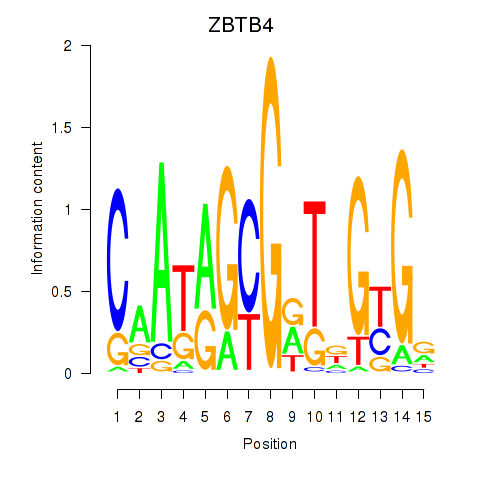
Results for ZBTB4
Z-value: 0.28
Transcription factors associated with ZBTB4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB4
|
ENSG00000174282.7 | ZBTB4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB4 | hg19_v2_chr17_-_7382834_7382855, hg19_v2_chr17_-_7387524_7387597 | -0.87 | 5.3e-03 | Click! |
Activity profile of ZBTB4 motif
Sorted Z-values of ZBTB4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_54671047 | 0.26 |
ENST00000332822.4 |
NOG |
noggin |
| chr16_-_87903079 | 0.24 |
ENST00000261622.4 |
SLC7A5 |
solute carrier family 7 (amino acid transporter light chain, L system), member 5 |
| chr11_+_62623544 | 0.23 |
ENST00000377890.2 ENST00000377891.2 ENST00000377889.2 |
SLC3A2 |
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr11_+_62623512 | 0.23 |
ENST00000377892.1 |
SLC3A2 |
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr5_-_150948414 | 0.20 |
ENST00000261800.5 |
FAT2 |
FAT atypical cadherin 2 |
| chr6_-_136847610 | 0.16 |
ENST00000454590.1 ENST00000432797.2 |
MAP7 |
microtubule-associated protein 7 |
| chr3_+_148583043 | 0.15 |
ENST00000296046.3 |
CPA3 |
carboxypeptidase A3 (mast cell) |
| chr3_+_113666748 | 0.11 |
ENST00000330212.3 ENST00000498275.1 |
ZDHHC23 |
zinc finger, DHHC-type containing 23 |
| chr22_+_29279552 | 0.11 |
ENST00000544604.2 |
ZNRF3 |
zinc and ring finger 3 |
| chr6_+_41606176 | 0.11 |
ENST00000441667.1 ENST00000230321.6 ENST00000373050.4 ENST00000446650.1 ENST00000435476.1 |
MDFI |
MyoD family inhibitor |
| chr15_+_90728145 | 0.11 |
ENST00000561085.1 ENST00000379122.3 ENST00000332496.6 |
SEMA4B |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr3_+_14989186 | 0.10 |
ENST00000435454.1 ENST00000323373.6 |
NR2C2 |
nuclear receptor subfamily 2, group C, member 2 |
| chr2_-_180871780 | 0.10 |
ENST00000410053.3 ENST00000295749.6 ENST00000404136.2 |
CWC22 |
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
| chr6_-_13487784 | 0.09 |
ENST00000379287.3 |
GFOD1 |
glucose-fructose oxidoreductase domain containing 1 |
| chr6_+_30882108 | 0.09 |
ENST00000541562.1 ENST00000421263.1 |
VARS2 |
valyl-tRNA synthetase 2, mitochondrial |
| chr2_+_192110199 | 0.09 |
ENST00000304164.4 |
MYO1B |
myosin IB |
| chr2_+_192109911 | 0.09 |
ENST00000418908.1 ENST00000339514.4 ENST00000392318.3 |
MYO1B |
myosin IB |
| chr19_+_39138320 | 0.08 |
ENST00000424234.2 ENST00000390009.3 ENST00000589528.1 |
ACTN4 |
actinin, alpha 4 |
| chr14_-_69445968 | 0.08 |
ENST00000438964.2 |
ACTN1 |
actinin, alpha 1 |
| chr19_-_5791215 | 0.08 |
ENST00000320699.8 ENST00000309061.7 |
DUS3L |
dihydrouridine synthase 3-like (S. cerevisiae) |
| chr22_-_20231207 | 0.08 |
ENST00000425986.1 |
RTN4R |
reticulon 4 receptor |
| chr11_+_65082289 | 0.08 |
ENST00000279249.2 |
CDC42EP2 |
CDC42 effector protein (Rho GTPase binding) 2 |
| chr13_+_108870714 | 0.08 |
ENST00000375898.3 |
ABHD13 |
abhydrolase domain containing 13 |
| chr10_-_101380121 | 0.08 |
ENST00000370495.4 |
SLC25A28 |
solute carrier family 25 (mitochondrial iron transporter), member 28 |
| chr12_+_106751436 | 0.08 |
ENST00000228347.4 |
POLR3B |
polymerase (RNA) III (DNA directed) polypeptide B |
| chr19_+_39421556 | 0.08 |
ENST00000407800.2 ENST00000402029.3 |
MRPS12 |
mitochondrial ribosomal protein S12 |
| chr19_+_10216899 | 0.07 |
ENST00000428358.1 ENST00000393796.4 ENST00000253107.7 ENST00000556468.1 ENST00000393793.1 |
PPAN-P2RY11 PPAN |
PPAN-P2RY11 readthrough peter pan homolog (Drosophila) |
| chr14_-_69445793 | 0.07 |
ENST00000538545.2 ENST00000394419.4 |
ACTN1 |
actinin, alpha 1 |
| chr8_+_94929077 | 0.07 |
ENST00000297598.4 ENST00000520614.1 |
PDP1 |
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr7_+_63774321 | 0.07 |
ENST00000423484.2 |
ZNF736 |
zinc finger protein 736 |
| chr14_+_23305760 | 0.06 |
ENST00000311852.6 |
MMP14 |
matrix metallopeptidase 14 (membrane-inserted) |
| chr19_+_39138271 | 0.06 |
ENST00000252699.2 |
ACTN4 |
actinin, alpha 4 |
| chr4_-_7069760 | 0.06 |
ENST00000264954.4 |
GRPEL1 |
GrpE-like 1, mitochondrial (E. coli) |
| chr4_+_128554081 | 0.06 |
ENST00000335251.6 ENST00000296461.5 |
INTU |
inturned planar cell polarity protein |
| chr11_-_63439381 | 0.06 |
ENST00000538786.1 ENST00000540699.1 |
ATL3 |
atlastin GTPase 3 |
| chr16_+_2285817 | 0.06 |
ENST00000564065.1 |
DNASE1L2 |
deoxyribonuclease I-like 2 |
| chr4_-_156297949 | 0.06 |
ENST00000515654.1 |
MAP9 |
microtubule-associated protein 9 |
| chr4_-_156298028 | 0.06 |
ENST00000433024.1 ENST00000379248.2 |
MAP9 |
microtubule-associated protein 9 |
| chr14_-_85996332 | 0.06 |
ENST00000380722.1 |
RP11-497E19.1 |
RP11-497E19.1 |
| chr1_-_156786530 | 0.06 |
ENST00000368198.3 |
SH2D2A |
SH2 domain containing 2A |
| chr9_-_131709858 | 0.06 |
ENST00000372586.3 |
DOLK |
dolichol kinase |
| chr16_-_89556942 | 0.06 |
ENST00000301030.4 |
ANKRD11 |
ankyrin repeat domain 11 |
| chr11_-_46940074 | 0.06 |
ENST00000378623.1 ENST00000534404.1 |
LRP4 |
low density lipoprotein receptor-related protein 4 |
| chr9_-_130497565 | 0.06 |
ENST00000336067.6 ENST00000373281.5 ENST00000373284.5 ENST00000458505.3 |
TOR2A |
torsin family 2, member A |
| chr8_-_74205851 | 0.06 |
ENST00000396467.1 |
RPL7 |
ribosomal protein L7 |
| chr13_+_24734880 | 0.06 |
ENST00000382095.4 |
SPATA13 |
spermatogenesis associated 13 |
| chr10_+_37414785 | 0.06 |
ENST00000374660.1 ENST00000602533.1 ENST00000361713.1 |
ANKRD30A |
ankyrin repeat domain 30A |
| chrX_+_100075368 | 0.06 |
ENST00000415585.2 ENST00000372972.2 ENST00000413437.1 |
CSTF2 |
cleavage stimulation factor, 3' pre-RNA, subunit 2, 64kDa |
| chr14_-_69446034 | 0.06 |
ENST00000193403.6 |
ACTN1 |
actinin, alpha 1 |
| chr9_-_93405352 | 0.06 |
ENST00000375765.3 |
DIRAS2 |
DIRAS family, GTP-binding RAS-like 2 |
| chr2_+_73612858 | 0.05 |
ENST00000409009.1 ENST00000264448.6 ENST00000377715.1 |
ALMS1 |
Alstrom syndrome 1 |
| chr14_+_85996507 | 0.05 |
ENST00000554746.1 |
FLRT2 |
fibronectin leucine rich transmembrane protein 2 |
| chr14_+_85996471 | 0.05 |
ENST00000330753.4 |
FLRT2 |
fibronectin leucine rich transmembrane protein 2 |
| chr22_+_32439019 | 0.05 |
ENST00000266088.4 |
SLC5A1 |
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr1_+_169075554 | 0.05 |
ENST00000367815.4 |
ATP1B1 |
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr8_-_145550571 | 0.05 |
ENST00000332324.4 |
DGAT1 |
diacylglycerol O-acyltransferase 1 |
| chr12_+_52281744 | 0.05 |
ENST00000301190.6 ENST00000538991.1 |
ANKRD33 |
ankyrin repeat domain 33 |
| chr4_-_156298087 | 0.05 |
ENST00000311277.4 |
MAP9 |
microtubule-associated protein 9 |
| chr8_+_94929168 | 0.05 |
ENST00000518107.1 ENST00000396200.3 |
PDP1 |
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr6_+_158244223 | 0.05 |
ENST00000392185.3 |
SNX9 |
sorting nexin 9 |
| chrX_+_147582130 | 0.05 |
ENST00000370460.2 ENST00000370457.5 |
AFF2 |
AF4/FMR2 family, member 2 |
| chr22_+_21987005 | 0.05 |
ENST00000607942.1 ENST00000425975.1 ENST00000292779.3 |
CCDC116 |
coiled-coil domain containing 116 |
| chr21_-_38445011 | 0.05 |
ENST00000464265.1 ENST00000399102.1 |
PIGP |
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr6_+_159290917 | 0.05 |
ENST00000367072.1 |
C6orf99 |
chromosome 6 open reading frame 99 |
| chr19_-_39421377 | 0.04 |
ENST00000430193.3 ENST00000600042.1 ENST00000221431.6 |
SARS2 |
seryl-tRNA synthetase 2, mitochondrial |
| chr12_+_52282001 | 0.04 |
ENST00000340970.4 |
ANKRD33 |
ankyrin repeat domain 33 |
| chr10_+_123922941 | 0.04 |
ENST00000360561.3 |
TACC2 |
transforming, acidic coiled-coil containing protein 2 |
| chr6_+_49431073 | 0.04 |
ENST00000335783.3 |
CENPQ |
centromere protein Q |
| chr10_-_32345305 | 0.04 |
ENST00000302418.4 |
KIF5B |
kinesin family member 5B |
| chr2_+_220094479 | 0.04 |
ENST00000323348.5 ENST00000453432.1 ENST00000409849.1 ENST00000416565.1 ENST00000410034.3 ENST00000447157.1 |
ANKZF1 |
ankyrin repeat and zinc finger domain containing 1 |
| chr1_+_33219592 | 0.04 |
ENST00000373481.3 |
KIAA1522 |
KIAA1522 |
| chr19_+_19144384 | 0.04 |
ENST00000392335.2 ENST00000535612.1 ENST00000537263.1 ENST00000540707.1 ENST00000541725.1 ENST00000269932.6 ENST00000546344.1 ENST00000540792.1 ENST00000536098.1 ENST00000541898.1 ENST00000543877.1 |
ARMC6 |
armadillo repeat containing 6 |
| chr3_-_128185811 | 0.04 |
ENST00000469083.1 |
DNAJB8 |
DnaJ (Hsp40) homolog, subfamily B, member 8 |
| chr7_-_44613494 | 0.04 |
ENST00000431640.1 ENST00000258772.5 |
DDX56 |
DEAD (Asp-Glu-Ala-Asp) box helicase 56 |
| chr15_-_59225758 | 0.04 |
ENST00000558486.1 ENST00000560682.1 ENST00000249736.7 ENST00000559880.1 ENST00000536328.1 |
SLTM |
SAFB-like, transcription modulator |
| chr13_-_24007815 | 0.04 |
ENST00000382298.3 |
SACS |
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chr3_-_192635943 | 0.04 |
ENST00000392452.2 |
MB21D2 |
Mab-21 domain containing 2 |
| chr4_+_4388805 | 0.04 |
ENST00000504171.1 |
NSG1 |
Homo sapiens neuron specific gene family member 1 (NSG1), transcript variant 3, mRNA. |
| chr1_-_33283754 | 0.04 |
ENST00000373477.4 |
YARS |
tyrosyl-tRNA synthetase |
| chr19_-_17356697 | 0.04 |
ENST00000291442.3 |
NR2F6 |
nuclear receptor subfamily 2, group F, member 6 |
| chr21_-_38445470 | 0.04 |
ENST00000399098.1 |
PIGP |
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr11_+_118754475 | 0.04 |
ENST00000292174.4 |
CXCR5 |
chemokine (C-X-C motif) receptor 5 |
| chr8_-_29208183 | 0.04 |
ENST00000240100.2 |
DUSP4 |
dual specificity phosphatase 4 |
| chr8_-_143851203 | 0.04 |
ENST00000521396.1 ENST00000317543.7 |
LYNX1 |
Ly6/neurotoxin 1 |
| chr14_-_55493763 | 0.04 |
ENST00000455555.1 ENST00000360586.3 ENST00000421192.1 ENST00000420358.2 |
WDHD1 |
WD repeat and HMG-box DNA binding protein 1 |
| chr7_-_100487280 | 0.04 |
ENST00000388761.2 |
UFSP1 |
UFM1-specific peptidase 1 (non-functional) |
| chr9_-_111696340 | 0.04 |
ENST00000374647.5 |
IKBKAP |
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase complex-associated protein |
| chr22_-_29784519 | 0.03 |
ENST00000357586.2 ENST00000356015.2 ENST00000432560.2 ENST00000317368.7 |
AP1B1 |
adaptor-related protein complex 1, beta 1 subunit |
| chr20_-_43438912 | 0.03 |
ENST00000541604.2 ENST00000372851.3 |
RIMS4 |
regulating synaptic membrane exocytosis 4 |
| chr15_-_98417780 | 0.03 |
ENST00000503874.3 |
LINC00923 |
long intergenic non-protein coding RNA 923 |
| chr12_+_5541267 | 0.03 |
ENST00000423158.3 |
NTF3 |
neurotrophin 3 |
| chr22_+_42229100 | 0.03 |
ENST00000361204.4 |
SREBF2 |
sterol regulatory element binding transcription factor 2 |
| chr11_-_72385437 | 0.03 |
ENST00000418754.2 ENST00000542969.2 ENST00000334456.5 |
PDE2A |
phosphodiesterase 2A, cGMP-stimulated |
| chr9_+_131709966 | 0.03 |
ENST00000372577.2 |
NUP188 |
nucleoporin 188kDa |
| chr22_+_50312316 | 0.03 |
ENST00000328268.4 |
CRELD2 |
cysteine-rich with EGF-like domains 2 |
| chr17_-_5389477 | 0.03 |
ENST00000572834.1 ENST00000570848.1 ENST00000571971.1 ENST00000158771.4 |
DERL2 |
derlin 2 |
| chr19_+_47759716 | 0.03 |
ENST00000221922.6 |
CCDC9 |
coiled-coil domain containing 9 |
| chr1_+_151254738 | 0.03 |
ENST00000336715.6 ENST00000324048.5 ENST00000368879.2 |
ZNF687 |
zinc finger protein 687 |
| chr19_+_17830051 | 0.03 |
ENST00000594625.1 ENST00000324096.4 ENST00000600186.1 ENST00000597735.1 |
MAP1S |
microtubule-associated protein 1S |
| chr2_-_10952922 | 0.03 |
ENST00000272227.3 |
PDIA6 |
protein disulfide isomerase family A, member 6 |
| chr16_-_30042580 | 0.03 |
ENST00000380495.4 |
FAM57B |
family with sequence similarity 57, member B |
| chr4_+_183370146 | 0.03 |
ENST00000510504.1 |
TENM3 |
teneurin transmembrane protein 3 |
| chr1_-_54872059 | 0.03 |
ENST00000371320.3 |
SSBP3 |
single stranded DNA binding protein 3 |
| chrX_-_74145273 | 0.03 |
ENST00000055682.6 |
KIAA2022 |
KIAA2022 |
| chr3_-_128186091 | 0.03 |
ENST00000319153.3 |
DNAJB8 |
DnaJ (Hsp40) homolog, subfamily B, member 8 |
| chr1_-_165738085 | 0.03 |
ENST00000464650.1 ENST00000392129.6 |
TMCO1 |
transmembrane and coiled-coil domains 1 |
| chr3_-_167452703 | 0.03 |
ENST00000497056.2 ENST00000473645.2 |
PDCD10 |
programmed cell death 10 |
| chr3_-_167452614 | 0.03 |
ENST00000392750.2 ENST00000464360.1 ENST00000492139.1 ENST00000471885.1 ENST00000470131.1 |
PDCD10 |
programmed cell death 10 |
| chr17_-_4046257 | 0.03 |
ENST00000381638.2 |
ZZEF1 |
zinc finger, ZZ-type with EF-hand domain 1 |
| chr1_+_110163709 | 0.03 |
ENST00000369840.2 ENST00000527846.1 |
AMPD2 |
adenosine monophosphate deaminase 2 |
| chr2_+_152266392 | 0.03 |
ENST00000444746.2 ENST00000453091.2 ENST00000428287.2 ENST00000433166.2 ENST00000420714.3 ENST00000243326.5 ENST00000414861.2 |
RIF1 |
RAP1 interacting factor homolog (yeast) |
| chr2_-_10952832 | 0.03 |
ENST00000540494.1 |
PDIA6 |
protein disulfide isomerase family A, member 6 |
| chr22_-_50970919 | 0.03 |
ENST00000329363.4 ENST00000437588.1 |
ODF3B |
outer dense fiber of sperm tails 3B |
| chr19_+_15121532 | 0.03 |
ENST00000292574.3 |
CCDC105 |
coiled-coil domain containing 105 |
| chr21_+_34638656 | 0.03 |
ENST00000290200.2 |
IL10RB |
interleukin 10 receptor, beta |
| chr1_+_10534944 | 0.03 |
ENST00000356607.4 ENST00000538836.1 ENST00000491661.2 |
PEX14 |
peroxisomal biogenesis factor 14 |
| chr3_-_119813264 | 0.03 |
ENST00000264235.8 |
GSK3B |
glycogen synthase kinase 3 beta |
| chr10_-_103599591 | 0.02 |
ENST00000348850.5 |
KCNIP2 |
Kv channel interacting protein 2 |
| chr1_-_203055129 | 0.02 |
ENST00000241651.4 |
MYOG |
myogenin (myogenic factor 4) |
| chr19_+_1275917 | 0.02 |
ENST00000469144.1 |
C19orf24 |
chromosome 19 open reading frame 24 |
| chr2_-_191399426 | 0.02 |
ENST00000409150.3 |
TMEM194B |
transmembrane protein 194B |
| chr2_+_74361599 | 0.02 |
ENST00000401851.1 |
MGC10955 |
MGC10955 |
| chr8_-_16859690 | 0.02 |
ENST00000180166.5 |
FGF20 |
fibroblast growth factor 20 |
| chr1_+_154947126 | 0.02 |
ENST00000368439.1 |
CKS1B |
CDC28 protein kinase regulatory subunit 1B |
| chr4_-_83295103 | 0.02 |
ENST00000313899.7 ENST00000352301.4 ENST00000509107.1 ENST00000353341.4 ENST00000541060.1 |
HNRNPD |
heterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) |
| chr8_+_94929110 | 0.02 |
ENST00000520728.1 |
PDP1 |
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr17_+_7487146 | 0.02 |
ENST00000396501.4 ENST00000584378.1 ENST00000423172.2 ENST00000579445.1 ENST00000585217.1 ENST00000581380.1 |
MPDU1 |
mannose-P-dolichol utilization defect 1 |
| chr17_+_45727204 | 0.02 |
ENST00000290158.4 |
KPNB1 |
karyopherin (importin) beta 1 |
| chr17_-_48943706 | 0.02 |
ENST00000499247.2 |
TOB1 |
transducer of ERBB2, 1 |
| chr22_+_50312379 | 0.02 |
ENST00000407217.3 ENST00000403427.3 |
CRELD2 |
cysteine-rich with EGF-like domains 2 |
| chr12_-_46766577 | 0.02 |
ENST00000256689.5 |
SLC38A2 |
solute carrier family 38, member 2 |
| chr1_+_154947148 | 0.02 |
ENST00000368436.1 ENST00000308987.5 |
CKS1B |
CDC28 protein kinase regulatory subunit 1B |
| chr11_-_64546202 | 0.02 |
ENST00000377390.3 ENST00000227503.9 ENST00000377394.3 ENST00000422298.2 ENST00000334944.5 |
SF1 |
splicing factor 1 |
| chr9_-_35650900 | 0.02 |
ENST00000259608.3 |
SIT1 |
signaling threshold regulating transmembrane adaptor 1 |
| chr9_-_136242956 | 0.02 |
ENST00000371989.3 ENST00000485435.2 |
SURF4 |
surfeit 4 |
| chr19_+_10531150 | 0.02 |
ENST00000352831.6 |
PDE4A |
phosphodiesterase 4A, cAMP-specific |
| chr7_+_100136811 | 0.02 |
ENST00000300176.4 ENST00000262935.4 |
AGFG2 |
ArfGAP with FG repeats 2 |
| chr1_+_33439268 | 0.02 |
ENST00000594612.1 |
FKSG48 |
FKSG48 |
| chr7_-_128694927 | 0.02 |
ENST00000471166.1 ENST00000265388.5 |
TNPO3 |
transportin 3 |
| chr5_-_96143796 | 0.02 |
ENST00000296754.3 |
ERAP1 |
endoplasmic reticulum aminopeptidase 1 |
| chr13_+_28712614 | 0.02 |
ENST00000380958.3 |
PAN3 |
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr19_+_10828724 | 0.02 |
ENST00000585892.1 ENST00000314646.5 ENST00000359692.6 |
DNM2 |
dynamin 2 |
| chr19_+_10828795 | 0.02 |
ENST00000389253.4 ENST00000355667.6 ENST00000408974.4 |
DNM2 |
dynamin 2 |
| chr9_-_71629010 | 0.02 |
ENST00000377276.2 |
PRKACG |
protein kinase, cAMP-dependent, catalytic, gamma |
| chr1_+_65886262 | 0.02 |
ENST00000371065.4 |
LEPROT |
leptin receptor overlapping transcript |
| chr2_-_241497390 | 0.02 |
ENST00000272972.3 ENST00000401804.1 ENST00000361678.4 ENST00000405523.3 |
ANKMY1 |
ankyrin repeat and MYND domain containing 1 |
| chr5_+_173416162 | 0.02 |
ENST00000340147.6 |
C5orf47 |
chromosome 5 open reading frame 47 |
| chr6_+_31582961 | 0.02 |
ENST00000376059.3 ENST00000337917.7 |
AIF1 |
allograft inflammatory factor 1 |
| chr9_+_4792971 | 0.02 |
ENST00000381732.3 ENST00000442869.1 |
RCL1 |
RNA terminal phosphate cyclase-like 1 |
| chr3_+_186915274 | 0.02 |
ENST00000312295.4 |
RTP1 |
receptor (chemosensory) transporter protein 1 |
| chr15_-_77363513 | 0.02 |
ENST00000267970.4 |
TSPAN3 |
tetraspanin 3 |
| chr6_+_31583761 | 0.02 |
ENST00000376049.4 |
AIF1 |
allograft inflammatory factor 1 |
| chr1_-_165738072 | 0.02 |
ENST00000481278.1 |
TMCO1 |
transmembrane and coiled-coil domains 1 |
| chr1_+_166808692 | 0.02 |
ENST00000367876.4 |
POGK |
pogo transposable element with KRAB domain |
| chr2_+_155554797 | 0.02 |
ENST00000295101.2 |
KCNJ3 |
potassium inwardly-rectifying channel, subfamily J, member 3 |
| chr1_-_67896009 | 0.02 |
ENST00000370990.5 |
SERBP1 |
SERPINE1 mRNA binding protein 1 |
| chr6_-_31864977 | 0.02 |
ENST00000395728.3 ENST00000375528.4 |
EHMT2 |
euchromatic histone-lysine N-methyltransferase 2 |
| chr17_-_2996290 | 0.02 |
ENST00000331459.1 |
OR1D2 |
olfactory receptor, family 1, subfamily D, member 2 |
| chr7_+_30811004 | 0.01 |
ENST00000265299.6 |
FAM188B |
family with sequence similarity 188, member B |
| chrX_-_68385274 | 0.01 |
ENST00000374584.3 ENST00000590146.1 |
PJA1 |
praja ring finger 1, E3 ubiquitin protein ligase |
| chr12_+_112204691 | 0.01 |
ENST00000416293.3 ENST00000261733.2 |
ALDH2 |
aldehyde dehydrogenase 2 family (mitochondrial) |
| chr16_-_75301886 | 0.01 |
ENST00000393422.2 |
BCAR1 |
breast cancer anti-estrogen resistance 1 |
| chrX_-_68385354 | 0.01 |
ENST00000361478.1 |
PJA1 |
praja ring finger 1, E3 ubiquitin protein ligase |
| chr18_-_76738843 | 0.01 |
ENST00000575722.1 ENST00000583511.1 |
RP11-849I19.1 |
RP11-849I19.1 |
| chr19_-_39881669 | 0.01 |
ENST00000221266.7 |
PAF1 |
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chr8_+_145691411 | 0.01 |
ENST00000301332.2 |
KIFC2 |
kinesin family member C2 |
| chr1_+_110754094 | 0.01 |
ENST00000369787.3 ENST00000413138.3 ENST00000438661.2 |
KCNC4 |
potassium voltage-gated channel, Shaw-related subfamily, member 4 |
| chr1_-_67896069 | 0.01 |
ENST00000370995.2 ENST00000361219.6 |
SERBP1 |
SERPINE1 mRNA binding protein 1 |
| chr4_+_71554196 | 0.01 |
ENST00000254803.2 |
UTP3 |
UTP3, small subunit (SSU) processome component, homolog (S. cerevisiae) |
| chr7_-_5463175 | 0.01 |
ENST00000399537.4 ENST00000430969.1 |
TNRC18 |
trinucleotide repeat containing 18 |
| chr16_+_640201 | 0.01 |
ENST00000563109.1 |
RAB40C |
RAB40C, member RAS oncogene family |
| chr1_-_108742957 | 0.01 |
ENST00000565488.1 |
SLC25A24 |
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr17_-_56595196 | 0.01 |
ENST00000579921.1 ENST00000579925.1 ENST00000323456.5 |
MTMR4 |
myotubularin related protein 4 |
| chr22_-_50970506 | 0.01 |
ENST00000428989.2 ENST00000403326.1 |
ODF3B |
outer dense fiber of sperm tails 3B |
| chr19_-_41903161 | 0.01 |
ENST00000602129.1 ENST00000593771.1 ENST00000596905.1 ENST00000221233.4 |
EXOSC5 |
exosome component 5 |
| chr2_+_182756701 | 0.01 |
ENST00000409001.1 |
SSFA2 |
sperm specific antigen 2 |
| chr3_+_188889737 | 0.01 |
ENST00000345063.3 |
TPRG1 |
tumor protein p63 regulated 1 |
| chr17_+_5389605 | 0.01 |
ENST00000576988.1 ENST00000576570.1 ENST00000573759.1 |
MIS12 |
MIS12 kinetochore complex component |
| chr14_+_74416989 | 0.01 |
ENST00000334571.2 ENST00000554920.1 |
COQ6 |
coenzyme Q6 monooxygenase |
| chr12_+_6561190 | 0.01 |
ENST00000544021.1 ENST00000266556.7 |
TAPBPL |
TAP binding protein-like |
| chr20_+_61147651 | 0.01 |
ENST00000370527.3 ENST00000370524.2 |
C20orf166 |
chromosome 20 open reading frame 166 |
| chr11_+_125462690 | 0.01 |
ENST00000392708.4 ENST00000529196.1 ENST00000531491.1 |
STT3A |
STT3A, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr15_+_91446157 | 0.01 |
ENST00000559717.1 |
MAN2A2 |
mannosidase, alpha, class 2A, member 2 |
| chr6_-_13711773 | 0.01 |
ENST00000011619.3 |
RANBP9 |
RAN binding protein 9 |
| chr1_+_151043070 | 0.01 |
ENST00000368918.3 ENST00000368917.1 |
GABPB2 |
GA binding protein transcription factor, beta subunit 2 |
| chr19_-_39881777 | 0.01 |
ENST00000595564.1 ENST00000221265.3 |
PAF1 |
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chr2_-_201936302 | 0.01 |
ENST00000453765.1 ENST00000452799.1 ENST00000446678.1 ENST00000418596.3 |
FAM126B |
family with sequence similarity 126, member B |
| chr7_+_102937869 | 0.01 |
ENST00000249269.4 ENST00000428154.1 ENST00000420236.2 |
PMPCB |
peptidase (mitochondrial processing) beta |
| chr15_+_90792760 | 0.01 |
ENST00000339615.5 ENST00000438251.1 |
TTLL13 |
tubulin tyrosine ligase-like family, member 13 |
| chr12_-_49525175 | 0.01 |
ENST00000336023.5 ENST00000550367.1 ENST00000552984.1 ENST00000547476.1 |
TUBA1B |
tubulin, alpha 1b |
| chr19_+_10131437 | 0.01 |
ENST00000587782.1 |
RDH8 |
retinol dehydrogenase 8 (all-trans) |
| chr7_-_18067478 | 0.01 |
ENST00000506618.2 |
PRPS1L1 |
phosphoribosyl pyrophosphate synthetase 1-like 1 |
| chr2_-_38829768 | 0.01 |
ENST00000378915.3 |
HNRNPLL |
heterogeneous nuclear ribonucleoprotein L-like |
| chr10_+_121485588 | 0.00 |
ENST00000361976.2 ENST00000369083.3 |
INPP5F |
inositol polyphosphate-5-phosphatase F |
| chrX_-_63425561 | 0.00 |
ENST00000374869.3 ENST00000330258.3 |
AMER1 |
APC membrane recruitment protein 1 |
| chr15_+_42696954 | 0.00 |
ENST00000337571.4 ENST00000569136.1 |
CAPN3 |
calpain 3, (p94) |
| chr4_-_113437328 | 0.00 |
ENST00000313341.3 |
NEUROG2 |
neurogenin 2 |
| chr2_-_207024233 | 0.00 |
ENST00000423725.1 ENST00000233190.6 |
NDUFS1 |
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr3_+_159943362 | 0.00 |
ENST00000326474.3 |
C3orf80 |
chromosome 3 open reading frame 80 |
| chr9_+_33290491 | 0.00 |
ENST00000379540.3 ENST00000379521.4 ENST00000318524.6 |
NFX1 |
nuclear transcription factor, X-box binding 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0060356 | leucine import(GO:0060356) |
| 0.0 | 0.1 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.0 | 0.3 | GO:0060300 | regulation of cytokine activity(GO:0060300) BMP signaling pathway involved in heart development(GO:0061312) |
| 0.0 | 0.1 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.0 | 0.1 | GO:2000051 | Wnt receptor catabolic process(GO:0038018) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.1 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.1 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.0 | 0.2 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 | 0.1 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.0 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.0 | 0.1 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.0 | 0.0 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.0 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.0 | 0.1 | GO:1990834 | response to odorant(GO:1990834) |
| 0.0 | 0.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.1 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.0 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.1 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.0 | 0.1 | GO:1903281 | protein transport into plasma membrane raft(GO:0044861) positive regulation of calcium:sodium antiporter activity(GO:1903281) positive regulation of potassium ion import(GO:1903288) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.2 | GO:0000235 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.0 | 0.1 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.1 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.0 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.0 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.0 | GO:0099609 | microtubule lateral binding(GO:0099609) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |