Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for ZBTB7A_ZBTB7C
Z-value: 0.38
Transcription factors associated with ZBTB7A_ZBTB7C
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
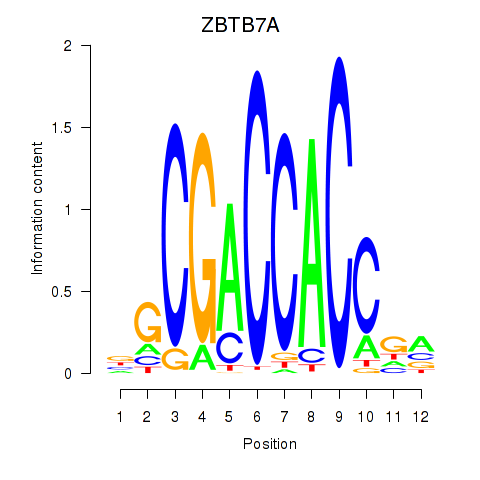
ZBTB7A
|
ENSG00000178951.4 | ZBTB7A |
|
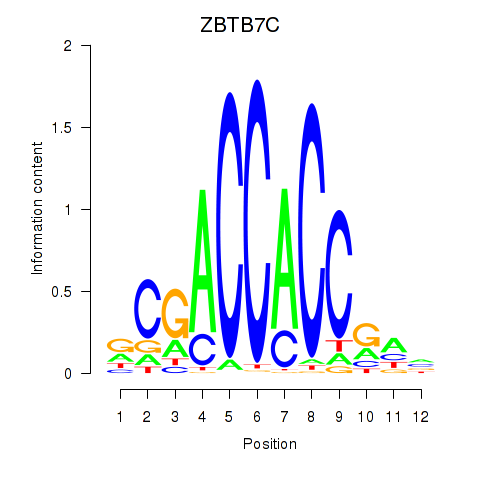
ZBTB7C
|
ENSG00000184828.5 | ZBTB7C |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB7A | hg19_v2_chr19_-_4066890_4066943 | -0.05 | 9.1e-01 | Click! |
| ZBTB7C | hg19_v2_chr18_-_45663666_45663732, hg19_v2_chr18_-_45935663_45935793 | 0.02 | 9.6e-01 | Click! |
Activity profile of ZBTB7A_ZBTB7C motif
Sorted Z-values of ZBTB7A_ZBTB7C motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB7A_ZBTB7C
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_235405168 | 0.41 |
ENST00000339728.3 |
ARL4C |
ADP-ribosylation factor-like 4C |
| chr5_+_149109825 | 0.19 |
ENST00000360453.4 ENST00000394320.3 ENST00000309241.5 |
PPARGC1B |
peroxisome proliferator-activated receptor gamma, coactivator 1 beta |
| chr6_+_73331520 | 0.14 |
ENST00000342056.2 ENST00000355194.4 |
KCNQ5 |
potassium voltage-gated channel, KQT-like subfamily, member 5 |
| chr1_-_38273840 | 0.14 |
ENST00000373044.2 |
YRDC |
yrdC N(6)-threonylcarbamoyltransferase domain containing |
| chr1_+_15480197 | 0.12 |
ENST00000400796.3 ENST00000434578.2 ENST00000376008.2 |
TMEM51 |
transmembrane protein 51 |
| chr1_-_92952433 | 0.12 |
ENST00000294702.5 |
GFI1 |
growth factor independent 1 transcription repressor |
| chr17_-_39674668 | 0.11 |
ENST00000393981.3 |
KRT15 |
keratin 15 |
| chr12_-_52867569 | 0.11 |
ENST00000252250.6 |
KRT6C |
keratin 6C |
| chr1_+_4714792 | 0.10 |
ENST00000378190.3 |
AJAP1 |
adherens junctions associated protein 1 |
| chr2_-_74692473 | 0.09 |
ENST00000535045.1 ENST00000409065.1 ENST00000414701.1 ENST00000448666.1 ENST00000233616.4 ENST00000452063.2 |
MOGS |
mannosyl-oligosaccharide glucosidase |
| chr19_+_38755203 | 0.09 |
ENST00000587090.1 ENST00000454580.3 |
SPINT2 |
serine peptidase inhibitor, Kunitz type, 2 |
| chr19_+_38755042 | 0.09 |
ENST00000301244.7 |
SPINT2 |
serine peptidase inhibitor, Kunitz type, 2 |
| chr7_+_26191809 | 0.09 |
ENST00000056233.3 |
NFE2L3 |
nuclear factor, erythroid 2-like 3 |
| chr12_-_52887034 | 0.09 |
ENST00000330722.6 |
KRT6A |
keratin 6A |
| chrX_+_17393543 | 0.08 |
ENST00000380060.3 |
NHS |
Nance-Horan syndrome (congenital cataracts and dental anomalies) |
| chr15_-_64126084 | 0.08 |
ENST00000560316.1 ENST00000443617.2 ENST00000560462.1 ENST00000558532.1 ENST00000561400.1 |
HERC1 |
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr1_-_95007193 | 0.08 |
ENST00000370207.4 ENST00000334047.7 |
F3 |
coagulation factor III (thromboplastin, tissue factor) |
| chr14_+_29236269 | 0.07 |
ENST00000313071.4 |
FOXG1 |
forkhead box G1 |
| chr19_+_35634146 | 0.07 |
ENST00000586063.1 ENST00000270310.2 ENST00000588265.1 |
FXYD7 |
FXYD domain containing ion transport regulator 7 |
| chr2_-_97405775 | 0.07 |
ENST00000264963.4 ENST00000537039.1 ENST00000377079.4 ENST00000426463.2 ENST00000534882.1 |
LMAN2L |
lectin, mannose-binding 2-like |
| chr5_+_137688285 | 0.07 |
ENST00000314358.5 |
KDM3B |
lysine (K)-specific demethylase 3B |
| chr11_-_17035943 | 0.07 |
ENST00000355661.3 ENST00000532079.1 ENST00000448080.2 ENST00000531066.1 |
PLEKHA7 |
pleckstrin homology domain containing, family A member 7 |
| chr7_-_94285511 | 0.06 |
ENST00000265735.7 |
SGCE |
sarcoglycan, epsilon |
| chrX_-_153141302 | 0.06 |
ENST00000361699.4 ENST00000543994.1 ENST00000370057.3 ENST00000538883.1 ENST00000361981.3 |
L1CAM |
L1 cell adhesion molecule |
| chrX_+_153991025 | 0.06 |
ENST00000369550.5 |
DKC1 |
dyskeratosis congenita 1, dyskerin |
| chr17_+_4736627 | 0.06 |
ENST00000355280.6 ENST00000347992.7 |
MINK1 |
misshapen-like kinase 1 |
| chr12_+_13197218 | 0.06 |
ENST00000197268.8 |
KIAA1467 |
KIAA1467 |
| chr11_-_62752162 | 0.05 |
ENST00000458333.2 ENST00000421062.2 |
SLC22A6 |
solute carrier family 22 (organic anion transporter), member 6 |
| chr5_+_72861560 | 0.05 |
ENST00000296792.4 ENST00000509005.1 ENST00000543251.1 ENST00000508686.1 ENST00000508491.1 |
UTP15 |
UTP15, U3 small nucleolar ribonucleoprotein, homolog (S. cerevisiae) |
| chr22_+_31031639 | 0.05 |
ENST00000343605.4 ENST00000300385.8 |
SLC35E4 |
solute carrier family 35, member E4 |
| chr1_+_169075554 | 0.05 |
ENST00000367815.4 |
ATP1B1 |
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr8_-_134584092 | 0.05 |
ENST00000522652.1 |
ST3GAL1 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr19_+_45394477 | 0.05 |
ENST00000252487.5 ENST00000405636.2 ENST00000592434.1 ENST00000426677.2 ENST00000589649.1 |
TOMM40 |
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr7_-_94285472 | 0.05 |
ENST00000437425.2 ENST00000447873.1 ENST00000415788.2 |
SGCE |
sarcoglycan, epsilon |
| chr6_+_125474992 | 0.05 |
ENST00000528193.1 |
TPD52L1 |
tumor protein D52-like 1 |
| chr9_+_74764278 | 0.05 |
ENST00000238018.4 ENST00000376989.3 |
GDA |
guanine deaminase |
| chr3_-_108308241 | 0.05 |
ENST00000295746.8 |
KIAA1524 |
KIAA1524 |
| chr6_+_125475335 | 0.05 |
ENST00000532429.1 ENST00000534199.1 |
TPD52L1 |
tumor protein D52-like 1 |
| chr1_+_45792541 | 0.05 |
ENST00000334815.3 |
HPDL |
4-hydroxyphenylpyruvate dioxygenase-like |
| chr19_-_49622348 | 0.05 |
ENST00000408991.2 |
C19orf73 |
chromosome 19 open reading frame 73 |
| chr14_-_102771516 | 0.04 |
ENST00000524214.1 ENST00000193029.6 ENST00000361847.2 |
MOK |
MOK protein kinase |
| chr16_+_53164833 | 0.04 |
ENST00000564845.1 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
| chr8_+_145202939 | 0.04 |
ENST00000423230.2 ENST00000398656.4 |
MROH1 |
maestro heat-like repeat family member 1 |
| chr11_-_129872712 | 0.04 |
ENST00000358825.5 ENST00000360871.3 ENST00000528746.1 |
PRDM10 |
PR domain containing 10 |
| chr10_-_105615164 | 0.04 |
ENST00000355946.2 ENST00000369774.4 |
SH3PXD2A |
SH3 and PX domains 2A |
| chr6_-_41715128 | 0.04 |
ENST00000356667.4 ENST00000373025.3 ENST00000425343.2 |
PGC |
progastricsin (pepsinogen C) |
| chr14_+_70346125 | 0.04 |
ENST00000361956.3 ENST00000381280.4 |
SMOC1 |
SPARC related modular calcium binding 1 |
| chr17_+_29158962 | 0.04 |
ENST00000321990.4 |
ATAD5 |
ATPase family, AAA domain containing 5 |
| chr3_-_33759699 | 0.04 |
ENST00000399362.4 ENST00000359576.5 ENST00000307312.7 |
CLASP2 |
cytoplasmic linker associated protein 2 |
| chr14_-_102771462 | 0.04 |
ENST00000522874.1 |
MOK |
MOK protein kinase |
| chr19_-_663277 | 0.04 |
ENST00000292363.5 |
RNF126 |
ring finger protein 126 |
| chr8_-_134584152 | 0.04 |
ENST00000521180.1 ENST00000517668.1 ENST00000319914.5 |
ST3GAL1 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr6_+_125474939 | 0.04 |
ENST00000527711.1 |
TPD52L1 |
tumor protein D52-like 1 |
| chr16_+_2521500 | 0.04 |
ENST00000293973.1 |
NTN3 |
netrin 3 |
| chr19_-_13044494 | 0.04 |
ENST00000593021.1 ENST00000587981.1 ENST00000423140.2 ENST00000314606.4 |
FARSA |
phenylalanyl-tRNA synthetase, alpha subunit |
| chr20_+_36149602 | 0.04 |
ENST00000062104.2 ENST00000346199.2 |
NNAT |
neuronatin |
| chrX_-_119603138 | 0.04 |
ENST00000200639.4 ENST00000371335.4 ENST00000538785.1 ENST00000434600.2 |
LAMP2 |
lysosomal-associated membrane protein 2 |
| chr17_+_61600682 | 0.04 |
ENST00000581784.1 ENST00000456941.2 ENST00000580652.1 ENST00000314672.5 ENST00000583023.1 |
KCNH6 |
potassium voltage-gated channel, subfamily H (eag-related), member 6 |
| chr19_+_4402659 | 0.04 |
ENST00000301280.5 ENST00000585854.1 |
CHAF1A |
chromatin assembly factor 1, subunit A (p150) |
| chr14_-_71276211 | 0.04 |
ENST00000381250.4 ENST00000555993.2 |
MAP3K9 |
mitogen-activated protein kinase kinase kinase 9 |
| chr17_-_39661849 | 0.04 |
ENST00000246635.3 ENST00000336861.3 ENST00000587544.1 ENST00000587435.1 |
KRT13 |
keratin 13 |
| chr3_-_33759541 | 0.04 |
ENST00000468888.2 |
CLASP2 |
cytoplasmic linker associated protein 2 |
| chr15_+_92396920 | 0.04 |
ENST00000318445.6 |
SLCO3A1 |
solute carrier organic anion transporter family, member 3A1 |
| chr7_-_94285402 | 0.04 |
ENST00000428696.2 ENST00000445866.2 |
SGCE |
sarcoglycan, epsilon |
| chr5_-_33984786 | 0.04 |
ENST00000296589.4 |
SLC45A2 |
solute carrier family 45, member 2 |
| chr20_-_17662705 | 0.04 |
ENST00000455029.2 |
RRBP1 |
ribosome binding protein 1 |
| chr10_+_101419187 | 0.04 |
ENST00000370489.4 |
ENTPD7 |
ectonucleoside triphosphate diphosphohydrolase 7 |
| chr4_-_83719884 | 0.04 |
ENST00000282709.4 ENST00000273908.4 |
SCD5 |
stearoyl-CoA desaturase 5 |
| chr7_+_116312411 | 0.04 |
ENST00000456159.1 ENST00000397752.3 ENST00000318493.6 |
MET |
met proto-oncogene |
| chr5_-_2751762 | 0.04 |
ENST00000302057.5 ENST00000382611.6 |
IRX2 |
iroquois homeobox 2 |
| chr5_-_33984741 | 0.04 |
ENST00000382102.3 ENST00000509381.1 ENST00000342059.3 ENST00000345083.5 |
SLC45A2 |
solute carrier family 45, member 2 |
| chr2_+_27346666 | 0.04 |
ENST00000316470.4 ENST00000416071.1 |
ABHD1 |
abhydrolase domain containing 1 |
| chr8_+_27629459 | 0.04 |
ENST00000523566.1 |
ESCO2 |
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr13_+_96204961 | 0.03 |
ENST00000299339.2 |
CLDN10 |
claudin 10 |
| chr12_-_56224546 | 0.03 |
ENST00000357606.3 ENST00000547445.1 |
DNAJC14 |
DnaJ (Hsp40) homolog, subfamily C, member 14 |
| chr14_+_59951161 | 0.03 |
ENST00000261247.9 ENST00000425728.2 ENST00000556985.1 ENST00000554271.1 ENST00000554795.1 |
JKAMP |
JNK1/MAPK8-associated membrane protein |
| chr8_-_144242020 | 0.03 |
ENST00000414417.2 |
LY6H |
lymphocyte antigen 6 complex, locus H |
| chr17_+_73201754 | 0.03 |
ENST00000583569.1 ENST00000245544.4 ENST00000579324.1 ENST00000541827.1 ENST00000579298.1 ENST00000447371.2 |
NUP85 |
nucleoporin 85kDa |
| chr3_+_111578640 | 0.03 |
ENST00000393925.3 |
PHLDB2 |
pleckstrin homology-like domain, family B, member 2 |
| chr3_+_47422485 | 0.03 |
ENST00000431726.1 ENST00000456221.1 ENST00000265562.4 |
PTPN23 |
protein tyrosine phosphatase, non-receptor type 23 |
| chr3_+_111578027 | 0.03 |
ENST00000431670.2 ENST00000412622.1 |
PHLDB2 |
pleckstrin homology-like domain, family B, member 2 |
| chr2_+_25016282 | 0.03 |
ENST00000260662.1 |
CENPO |
centromere protein O |
| chrX_-_128977364 | 0.03 |
ENST00000371064.3 |
ZDHHC9 |
zinc finger, DHHC-type containing 9 |
| chr5_-_114515734 | 0.03 |
ENST00000514154.1 ENST00000282369.3 |
TRIM36 |
tripartite motif containing 36 |
| chr22_+_41487711 | 0.03 |
ENST00000263253.7 |
EP300 |
E1A binding protein p300 |
| chr2_-_37311445 | 0.03 |
ENST00000233099.5 ENST00000354531.2 |
HEATR5B |
HEAT repeat containing 5B |
| chr19_+_49622646 | 0.03 |
ENST00000334186.4 |
PPFIA3 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr1_+_167691191 | 0.03 |
ENST00000392121.3 ENST00000474859.1 |
MPZL1 |
myelin protein zero-like 1 |
| chr8_-_144241664 | 0.03 |
ENST00000342752.4 |
LY6H |
lymphocyte antigen 6 complex, locus H |
| chr16_-_19896220 | 0.03 |
ENST00000562469.1 ENST00000300571.2 |
GPRC5B |
G protein-coupled receptor, family C, group 5, member B |
| chr19_-_5567996 | 0.03 |
ENST00000448587.1 |
TINCR |
tissue differentiation-inducing non-protein coding RNA |
| chr15_-_41408409 | 0.03 |
ENST00000361937.3 |
INO80 |
INO80 complex subunit |
| chr15_-_41408339 | 0.03 |
ENST00000401393.3 |
INO80 |
INO80 complex subunit |
| chrX_+_152783131 | 0.03 |
ENST00000349466.2 ENST00000370186.1 |
ATP2B3 |
ATPase, Ca++ transporting, plasma membrane 3 |
| chr1_+_167691185 | 0.03 |
ENST00000359523.2 |
MPZL1 |
myelin protein zero-like 1 |
| chr7_-_150780487 | 0.03 |
ENST00000482202.1 |
TMUB1 |
transmembrane and ubiquitin-like domain containing 1 |
| chr2_+_25015968 | 0.03 |
ENST00000380834.2 ENST00000473706.1 |
CENPO |
centromere protein O |
| chr8_+_73449625 | 0.03 |
ENST00000523207.1 |
KCNB2 |
potassium voltage-gated channel, Shab-related subfamily, member 2 |
| chr4_-_7069760 | 0.03 |
ENST00000264954.4 |
GRPEL1 |
GrpE-like 1, mitochondrial (E. coli) |
| chr7_+_77167376 | 0.03 |
ENST00000435495.2 |
PTPN12 |
protein tyrosine phosphatase, non-receptor type 12 |
| chr19_+_36036583 | 0.03 |
ENST00000392205.1 |
TMEM147 |
transmembrane protein 147 |
| chr16_-_49315731 | 0.03 |
ENST00000219197.6 |
CBLN1 |
cerebellin 1 precursor |
| chr11_-_57103327 | 0.03 |
ENST00000529002.1 ENST00000278412.2 |
SSRP1 |
structure specific recognition protein 1 |
| chr1_-_111506562 | 0.03 |
ENST00000485275.2 ENST00000369763.4 |
LRIF1 |
ligand dependent nuclear receptor interacting factor 1 |
| chr14_-_51411146 | 0.03 |
ENST00000532462.1 |
PYGL |
phosphorylase, glycogen, liver |
| chr5_+_38258511 | 0.03 |
ENST00000354891.3 ENST00000322350.5 |
EGFLAM |
EGF-like, fibronectin type III and laminin G domains |
| chr19_+_39833036 | 0.03 |
ENST00000602243.1 ENST00000598913.1 ENST00000314471.6 |
SAMD4B |
sterile alpha motif domain containing 4B |
| chr3_+_111578131 | 0.03 |
ENST00000498699.1 |
PHLDB2 |
pleckstrin homology-like domain, family B, member 2 |
| chr14_+_59655369 | 0.03 |
ENST00000360909.3 ENST00000351081.1 ENST00000556135.1 |
DAAM1 |
dishevelled associated activator of morphogenesis 1 |
| chr8_-_144241432 | 0.03 |
ENST00000430474.2 |
LY6H |
lymphocyte antigen 6 complex, locus H |
| chr17_-_58469591 | 0.03 |
ENST00000589335.1 |
USP32 |
ubiquitin specific peptidase 32 |
| chr20_-_61557821 | 0.03 |
ENST00000354665.4 ENST00000370368.1 ENST00000395343.1 ENST00000395340.1 |
DIDO1 |
death inducer-obliterator 1 |
| chr17_-_46799872 | 0.03 |
ENST00000290294.3 |
PRAC1 |
prostate cancer susceptibility candidate 1 |
| chr19_+_36036477 | 0.02 |
ENST00000222284.5 ENST00000392204.2 |
TMEM147 |
transmembrane protein 147 |
| chr17_-_5372271 | 0.02 |
ENST00000225296.3 |
DHX33 |
DEAH (Asp-Glu-Ala-His) box polypeptide 33 |
| chr14_-_51411194 | 0.02 |
ENST00000544180.2 |
PYGL |
phosphorylase, glycogen, liver |
| chr6_-_166755995 | 0.02 |
ENST00000361731.3 |
SFT2D1 |
SFT2 domain containing 1 |
| chr19_-_45873642 | 0.02 |
ENST00000485403.2 ENST00000586856.1 ENST00000586131.1 ENST00000391940.4 ENST00000221481.6 ENST00000391944.3 ENST00000391945.4 |
ERCC2 |
excision repair cross-complementing rodent repair deficiency, complementation group 2 |
| chr7_-_112430427 | 0.02 |
ENST00000449743.1 ENST00000441474.1 ENST00000454074.1 ENST00000447395.1 |
TMEM168 |
transmembrane protein 168 |
| chr9_+_109625378 | 0.02 |
ENST00000277225.5 ENST00000457913.1 ENST00000472574.1 |
ZNF462 |
zinc finger protein 462 |
| chr16_+_84002234 | 0.02 |
ENST00000305202.4 |
NECAB2 |
N-terminal EF-hand calcium binding protein 2 |
| chrX_-_51812268 | 0.02 |
ENST00000486010.1 ENST00000497164.1 ENST00000360134.6 ENST00000485287.1 ENST00000335504.5 ENST00000431659.1 |
MAGED4B |
melanoma antigen family D, 4B |
| chr10_-_101769617 | 0.02 |
ENST00000324109.4 ENST00000342239.3 |
DNMBP |
dynamin binding protein |
| chr10_-_131762105 | 0.02 |
ENST00000368648.3 ENST00000355311.5 |
EBF3 |
early B-cell factor 3 |
| chr9_-_77643307 | 0.02 |
ENST00000376834.3 ENST00000376830.3 |
C9orf41 |
chromosome 9 open reading frame 41 |
| chr20_+_35201993 | 0.02 |
ENST00000373872.4 |
TGIF2 |
TGFB-induced factor homeobox 2 |
| chr14_-_23904861 | 0.02 |
ENST00000355349.3 |
MYH7 |
myosin, heavy chain 7, cardiac muscle, beta |
| chr15_+_92397051 | 0.02 |
ENST00000424469.2 |
SLCO3A1 |
solute carrier organic anion transporter family, member 3A1 |
| chr1_+_110754094 | 0.02 |
ENST00000369787.3 ENST00000413138.3 ENST00000438661.2 |
KCNC4 |
potassium voltage-gated channel, Shaw-related subfamily, member 4 |
| chr9_+_74764340 | 0.02 |
ENST00000376986.1 ENST00000358399.3 |
GDA |
guanine deaminase |
| chr21_-_47648665 | 0.02 |
ENST00000450351.1 ENST00000522411.1 ENST00000356396.4 ENST00000397728.3 ENST00000457828.2 |
LSS |
lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) |
| chr14_+_39644387 | 0.02 |
ENST00000553331.1 ENST00000216832.4 |
PNN |
pinin, desmosome associated protein |
| chr12_+_132312931 | 0.02 |
ENST00000360564.1 ENST00000545671.1 ENST00000545790.1 |
MMP17 |
matrix metallopeptidase 17 (membrane-inserted) |
| chr20_-_23402028 | 0.02 |
ENST00000398425.3 ENST00000432543.2 ENST00000377026.4 |
NAPB |
N-ethylmaleimide-sensitive factor attachment protein, beta |
| chr1_+_89246647 | 0.02 |
ENST00000544045.1 |
PKN2 |
protein kinase N2 |
| chrX_-_128977781 | 0.02 |
ENST00000357166.6 |
ZDHHC9 |
zinc finger, DHHC-type containing 9 |
| chr2_+_27505260 | 0.02 |
ENST00000380075.2 ENST00000296098.4 |
TRIM54 |
tripartite motif containing 54 |
| chr17_-_39769005 | 0.02 |
ENST00000301653.4 ENST00000593067.1 |
KRT16 |
keratin 16 |
| chr11_-_67771513 | 0.02 |
ENST00000227471.2 |
UNC93B1 |
unc-93 homolog B1 (C. elegans) |
| chr19_+_40697514 | 0.02 |
ENST00000253055.3 |
MAP3K10 |
mitogen-activated protein kinase kinase kinase 10 |
| chr9_-_96717654 | 0.02 |
ENST00000253968.6 |
BARX1 |
BARX homeobox 1 |
| chr11_-_62752455 | 0.02 |
ENST00000360421.4 |
SLC22A6 |
solute carrier family 22 (organic anion transporter), member 6 |
| chr6_+_87865262 | 0.02 |
ENST00000369577.3 ENST00000518845.1 ENST00000339907.4 ENST00000496806.2 |
ZNF292 |
zinc finger protein 292 |
| chr16_+_215965 | 0.02 |
ENST00000356815.3 |
HBM |
hemoglobin, mu |
| chr8_+_75896731 | 0.02 |
ENST00000262207.4 |
CRISPLD1 |
cysteine-rich secretory protein LCCL domain containing 1 |
| chrX_-_122866874 | 0.02 |
ENST00000245838.8 ENST00000355725.4 |
THOC2 |
THO complex 2 |
| chr11_-_62752429 | 0.02 |
ENST00000377871.3 |
SLC22A6 |
solute carrier family 22 (organic anion transporter), member 6 |
| chr13_+_110959598 | 0.02 |
ENST00000360467.5 |
COL4A2 |
collagen, type IV, alpha 2 |
| chr22_+_38035459 | 0.02 |
ENST00000357436.4 |
SH3BP1 |
SH3-domain binding protein 1 |
| chr2_+_58655461 | 0.02 |
ENST00000429095.1 ENST00000429664.1 ENST00000452840.1 |
AC007092.1 |
long intergenic non-protein coding RNA 1122 |
| chr7_-_100034060 | 0.02 |
ENST00000292330.2 |
PPP1R35 |
protein phosphatase 1, regulatory subunit 35 |
| chr1_+_25664408 | 0.02 |
ENST00000374358.4 |
TMEM50A |
transmembrane protein 50A |
| chr14_-_24584138 | 0.02 |
ENST00000558280.1 ENST00000561028.1 |
NRL |
neural retina leucine zipper |
| chr12_-_107487604 | 0.02 |
ENST00000008527.5 |
CRY1 |
cryptochrome 1 (photolyase-like) |
| chr22_+_42229100 | 0.02 |
ENST00000361204.4 |
SREBF2 |
sterol regulatory element binding transcription factor 2 |
| chr7_-_150779995 | 0.02 |
ENST00000462940.1 ENST00000492838.1 ENST00000392818.3 ENST00000488752.1 ENST00000476627.1 |
TMUB1 |
transmembrane and ubiquitin-like domain containing 1 |
| chr21_+_45079409 | 0.02 |
ENST00000340648.4 |
RRP1B |
ribosomal RNA processing 1B |
| chr7_-_150780609 | 0.02 |
ENST00000297533.4 |
TMUB1 |
transmembrane and ubiquitin-like domain containing 1 |
| chr7_-_151107106 | 0.02 |
ENST00000334493.6 |
WDR86 |
WD repeat domain 86 |
| chr20_-_36156293 | 0.02 |
ENST00000373537.2 ENST00000414542.2 |
BLCAP |
bladder cancer associated protein |
| chr11_+_133938955 | 0.02 |
ENST00000534549.1 ENST00000441717.3 |
JAM3 |
junctional adhesion molecule 3 |
| chr15_+_76352178 | 0.02 |
ENST00000388942.3 |
C15orf27 |
chromosome 15 open reading frame 27 |
| chr4_+_72053017 | 0.02 |
ENST00000351898.6 |
SLC4A4 |
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chr14_-_68283291 | 0.02 |
ENST00000555452.1 ENST00000347230.4 |
ZFYVE26 |
zinc finger, FYVE domain containing 26 |
| chr16_+_2014993 | 0.02 |
ENST00000564014.1 |
SNHG9 |
small nucleolar RNA host gene 9 (non-protein coding) |
| chr20_-_36156125 | 0.02 |
ENST00000397135.1 ENST00000397137.1 |
BLCAP |
bladder cancer associated protein |
| chr1_-_201123586 | 0.02 |
ENST00000414605.2 ENST00000367334.5 ENST00000367332.1 |
TMEM9 |
transmembrane protein 9 |
| chr11_+_133938820 | 0.02 |
ENST00000299106.4 ENST00000529443.2 |
JAM3 |
junctional adhesion molecule 3 |
| chr3_+_179040767 | 0.01 |
ENST00000496856.1 ENST00000491818.1 |
ZNF639 |
zinc finger protein 639 |
| chr6_-_109703634 | 0.01 |
ENST00000324953.5 ENST00000310786.4 ENST00000275080.7 ENST00000413644.2 |
CD164 |
CD164 molecule, sialomucin |
| chr5_-_139283982 | 0.01 |
ENST00000340391.3 |
NRG2 |
neuregulin 2 |
| chr1_-_201123546 | 0.01 |
ENST00000435310.1 ENST00000485839.2 ENST00000367330.1 |
TMEM9 |
transmembrane protein 9 |
| chr7_+_66093908 | 0.01 |
ENST00000443322.1 ENST00000449064.1 |
KCTD7 |
potassium channel tetramerization domain containing 7 |
| chr14_+_29234870 | 0.01 |
ENST00000382535.3 |
FOXG1 |
forkhead box G1 |
| chr10_+_60145155 | 0.01 |
ENST00000373895.3 |
TFAM |
transcription factor A, mitochondrial |
| chr10_+_14880157 | 0.01 |
ENST00000378372.3 |
HSPA14 |
heat shock 70kDa protein 14 |
| chr12_-_106641728 | 0.01 |
ENST00000378026.4 |
CKAP4 |
cytoskeleton-associated protein 4 |
| chr1_-_43205811 | 0.01 |
ENST00000372539.3 ENST00000296387.1 ENST00000539749.1 |
CLDN19 |
claudin 19 |
| chr1_+_19991780 | 0.01 |
ENST00000289753.1 |
HTR6 |
5-hydroxytryptamine (serotonin) receptor 6, G protein-coupled |
| chr1_-_111217603 | 0.01 |
ENST00000369769.2 |
KCNA3 |
potassium voltage-gated channel, shaker-related subfamily, member 3 |
| chr3_+_15468862 | 0.01 |
ENST00000396842.2 |
EAF1 |
ELL associated factor 1 |
| chr7_+_77167343 | 0.01 |
ENST00000433369.2 ENST00000415482.2 |
PTPN12 |
protein tyrosine phosphatase, non-receptor type 12 |
| chr4_-_76598326 | 0.01 |
ENST00000503660.1 |
G3BP2 |
GTPase activating protein (SH3 domain) binding protein 2 |
| chr1_-_151431909 | 0.01 |
ENST00000361398.3 ENST00000271715.2 |
POGZ |
pogo transposable element with ZNF domain |
| chr5_-_145214848 | 0.01 |
ENST00000505416.1 ENST00000334744.4 ENST00000358004.2 ENST00000511435.1 |
PRELID2 |
PRELI domain containing 2 |
| chrX_+_133507283 | 0.01 |
ENST00000370803.3 |
PHF6 |
PHD finger protein 6 |
| chr6_-_30043539 | 0.01 |
ENST00000376751.3 ENST00000244360.6 |
RNF39 |
ring finger protein 39 |
| chr4_+_72052964 | 0.01 |
ENST00000264485.5 ENST00000425175.1 |
SLC4A4 |
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chr9_+_140500087 | 0.01 |
ENST00000371421.4 |
ARRDC1 |
arrestin domain containing 1 |
| chr12_+_22199108 | 0.01 |
ENST00000229329.2 |
CMAS |
cytidine monophosphate N-acetylneuraminic acid synthetase |
| chr5_-_140070897 | 0.01 |
ENST00000448240.1 ENST00000438307.2 ENST00000415192.2 ENST00000457527.2 ENST00000307633.3 ENST00000507746.1 ENST00000431330.2 |
HARS |
histidyl-tRNA synthetase |
| chr16_+_12995614 | 0.01 |
ENST00000423335.2 |
SHISA9 |
shisa family member 9 |
| chr6_-_17706618 | 0.01 |
ENST00000262077.2 ENST00000537253.1 |
NUP153 |
nucleoporin 153kDa |
| chr15_-_62352659 | 0.01 |
ENST00000249837.3 |
VPS13C |
vacuolar protein sorting 13 homolog C (S. cerevisiae) |
| chr17_-_58469474 | 0.01 |
ENST00000300896.4 |
USP32 |
ubiquitin specific peptidase 32 |
| chr15_+_90895471 | 0.01 |
ENST00000354377.3 ENST00000379090.5 |
ZNF774 |
zinc finger protein 774 |
| chr6_-_122792919 | 0.01 |
ENST00000339697.4 |
SERINC1 |
serine incorporator 1 |
| chr20_-_35492048 | 0.01 |
ENST00000237536.4 |
SOGA1 |
suppressor of glucose, autophagy associated 1 |
| chr10_+_14880287 | 0.01 |
ENST00000437161.2 |
HSPA14 |
heat shock 70kDa protein 14 |
| chr5_-_145214893 | 0.01 |
ENST00000394450.2 |
PRELID2 |
PRELI domain containing 2 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.2 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.1 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.0 | 0.1 | GO:0097254 | renal tubular secretion(GO:0097254) |
| 0.0 | 0.1 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.1 | GO:0015766 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.2 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.1 | GO:0051710 | cytolysis by symbiont of host cells(GO:0001897) regulation of cytolysis in other organism(GO:0051710) |
| 0.0 | 0.1 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.0 | 0.0 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.1 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.0 | 0.0 | GO:0045799 | positive regulation of chromatin assembly or disassembly(GO:0045799) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.1 | GO:1903288 | protein transport into plasma membrane raft(GO:0044861) positive regulation of calcium:sodium antiporter activity(GO:1903281) positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.1 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0090661 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0008506 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0008184 | purine nucleobase binding(GO:0002060) glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |