Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
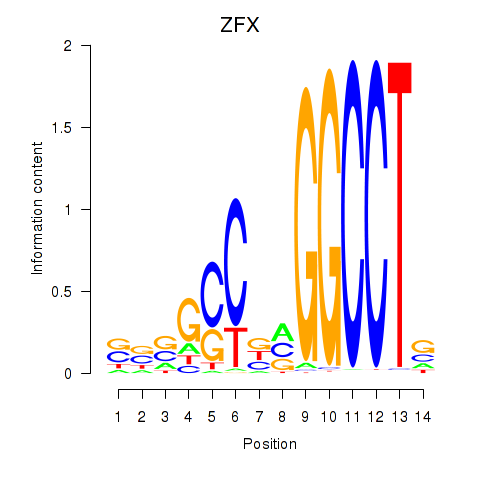
Results for ZFX
Z-value: 1.24
Transcription factors associated with ZFX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZFX
|
ENSG00000005889.11 | ZFX |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZFX | hg19_v2_chrX_+_24167746_24167811 | 0.85 | 7.8e-03 | Click! |
Activity profile of ZFX motif
Sorted Z-values of ZFX motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZFX
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_176560007 | 0.57 |
ENST00000510954.1 ENST00000354179.4 |
NSD1 |
nuclear receptor binding SET domain protein 1 |
| chr5_+_176560742 | 0.52 |
ENST00000439151.2 |
NSD1 |
nuclear receptor binding SET domain protein 1 |
| chr20_+_55204351 | 0.40 |
ENST00000201031.2 |
TFAP2C |
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr16_+_67063262 | 0.36 |
ENST00000565389.1 |
CBFB |
core-binding factor, beta subunit |
| chr1_-_11714700 | 0.36 |
ENST00000354287.4 |
FBXO2 |
F-box protein 2 |
| chr19_+_45844032 | 0.33 |
ENST00000589837.1 |
KLC3 |
kinesin light chain 3 |
| chr16_+_67062996 | 0.32 |
ENST00000561924.2 |
CBFB |
core-binding factor, beta subunit |
| chrX_+_24483338 | 0.29 |
ENST00000379162.4 ENST00000441463.2 |
PDK3 |
pyruvate dehydrogenase kinase, isozyme 3 |
| chr7_-_76256557 | 0.28 |
ENST00000275569.4 ENST00000310842.4 |
POMZP3 |
POM121 and ZP3 fusion |
| chr14_+_104552016 | 0.28 |
ENST00000551177.1 ENST00000546892.2 ENST00000455920.2 |
ASPG |
asparaginase homolog (S. cerevisiae) |
| chr20_-_30310797 | 0.28 |
ENST00000422920.1 |
BCL2L1 |
BCL2-like 1 |
| chr19_+_1065922 | 0.28 |
ENST00000539243.2 |
HMHA1 |
histocompatibility (minor) HA-1 |
| chr19_+_45843994 | 0.28 |
ENST00000391946.2 |
KLC3 |
kinesin light chain 3 |
| chr19_+_45844018 | 0.27 |
ENST00000585434.1 |
KLC3 |
kinesin light chain 3 |
| chr22_-_37823468 | 0.27 |
ENST00000402918.2 |
ELFN2 |
extracellular leucine-rich repeat and fibronectin type III domain containing 2 |
| chr16_+_765092 | 0.25 |
ENST00000568223.2 |
METRN |
meteorin, glial cell differentiation regulator |
| chr8_+_81398444 | 0.25 |
ENST00000455036.3 ENST00000426744.2 |
ZBTB10 |
zinc finger and BTB domain containing 10 |
| chr1_-_1850697 | 0.25 |
ENST00000378598.4 ENST00000416272.1 ENST00000310991.3 |
TMEM52 |
transmembrane protein 52 |
| chr4_+_1873100 | 0.24 |
ENST00000508803.1 |
WHSC1 |
Wolf-Hirschhorn syndrome candidate 1 |
| chr12_-_56882136 | 0.23 |
ENST00000311966.4 |
GLS2 |
glutaminase 2 (liver, mitochondrial) |
| chr20_-_1373682 | 0.23 |
ENST00000381724.3 |
FKBP1A |
FK506 binding protein 1A, 12kDa |
| chr7_-_44365020 | 0.23 |
ENST00000395747.2 ENST00000347193.4 ENST00000346990.4 ENST00000258682.6 ENST00000353625.4 ENST00000421607.1 ENST00000424197.1 ENST00000502837.2 ENST00000350811.3 ENST00000395749.2 |
CAMK2B |
calcium/calmodulin-dependent protein kinase II beta |
| chr14_-_105262016 | 0.23 |
ENST00000407796.2 |
AKT1 |
v-akt murine thymoma viral oncogene homolog 1 |
| chr19_+_39390320 | 0.23 |
ENST00000576510.1 |
NFKBIB |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr14_-_105262055 | 0.23 |
ENST00000349310.3 |
AKT1 |
v-akt murine thymoma viral oncogene homolog 1 |
| chr6_+_37137939 | 0.23 |
ENST00000373509.5 |
PIM1 |
pim-1 oncogene |
| chr16_-_57831914 | 0.23 |
ENST00000421376.2 |
KIFC3 |
kinesin family member C3 |
| chr14_+_74058410 | 0.22 |
ENST00000326303.4 |
ACOT4 |
acyl-CoA thioesterase 4 |
| chr16_+_67562702 | 0.22 |
ENST00000379312.3 ENST00000042381.4 ENST00000540839.3 |
FAM65A |
family with sequence similarity 65, member A |
| chr1_-_33430286 | 0.22 |
ENST00000373456.7 ENST00000356990.5 ENST00000235150.4 |
RNF19B |
ring finger protein 19B |
| chr12_+_77158021 | 0.21 |
ENST00000550876.1 |
ZDHHC17 |
zinc finger, DHHC-type containing 17 |
| chr11_+_71498552 | 0.21 |
ENST00000346333.6 ENST00000359244.4 ENST00000426628.2 |
FAM86C1 |
family with sequence similarity 86, member C1 |
| chr11_-_64570706 | 0.21 |
ENST00000294066.2 ENST00000377350.3 |
MAP4K2 |
mitogen-activated protein kinase kinase kinase kinase 2 |
| chr19_-_55658687 | 0.21 |
ENST00000593046.1 |
TNNT1 |
troponin T type 1 (skeletal, slow) |
| chr3_-_52090461 | 0.21 |
ENST00000296483.6 ENST00000495880.1 |
DUSP7 |
dual specificity phosphatase 7 |
| chr2_+_64681103 | 0.21 |
ENST00000464281.1 |
LGALSL |
lectin, galactoside-binding-like |
| chr11_+_128563652 | 0.20 |
ENST00000527786.2 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr10_-_120355149 | 0.20 |
ENST00000239032.2 |
PRLHR |
prolactin releasing hormone receptor |
| chr16_+_84682108 | 0.20 |
ENST00000564996.1 ENST00000258157.5 ENST00000567410.1 |
KLHL36 |
kelch-like family member 36 |
| chr10_+_134351319 | 0.20 |
ENST00000368594.3 ENST00000368593.3 |
INPP5A |
inositol polyphosphate-5-phosphatase, 40kDa |
| chr11_+_128563948 | 0.20 |
ENST00000534087.2 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr1_+_44444865 | 0.20 |
ENST00000372324.1 |
B4GALT2 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr8_-_143859197 | 0.19 |
ENST00000395192.2 |
LYNX1 |
Ly6/neurotoxin 1 |
| chr9_+_95820966 | 0.19 |
ENST00000375472.3 ENST00000465709.1 |
SUSD3 |
sushi domain containing 3 |
| chr11_+_66824276 | 0.19 |
ENST00000308831.2 |
RHOD |
ras homolog family member D |
| chr20_-_30310656 | 0.19 |
ENST00000376055.4 |
BCL2L1 |
BCL2-like 1 |
| chr19_+_51226648 | 0.19 |
ENST00000599973.1 |
CLEC11A |
C-type lectin domain family 11, member A |
| chr19_+_18942720 | 0.19 |
ENST00000262803.5 |
UPF1 |
UPF1 regulator of nonsense transcripts homolog (yeast) |
| chr20_-_34042558 | 0.19 |
ENST00000374372.1 |
GDF5 |
growth differentiation factor 5 |
| chr14_+_29234870 | 0.19 |
ENST00000382535.3 |
FOXG1 |
forkhead box G1 |
| chr19_-_55658650 | 0.19 |
ENST00000589226.1 |
TNNT1 |
troponin T type 1 (skeletal, slow) |
| chrX_+_150151752 | 0.19 |
ENST00000325307.7 |
HMGB3 |
high mobility group box 3 |
| chr3_-_71803474 | 0.18 |
ENST00000448225.1 ENST00000496214.2 |
EIF4E3 |
eukaryotic translation initiation factor 4E family member 3 |
| chr16_+_67063855 | 0.18 |
ENST00000563939.2 |
CBFB |
core-binding factor, beta subunit |
| chr14_+_105886150 | 0.18 |
ENST00000331320.7 ENST00000406191.1 |
MTA1 |
metastasis associated 1 |
| chr9_-_136004782 | 0.18 |
ENST00000393157.3 |
RALGDS |
ral guanine nucleotide dissociation stimulator |
| chr19_+_4343584 | 0.18 |
ENST00000596722.1 |
MPND |
MPN domain containing |
| chr2_-_165697920 | 0.18 |
ENST00000342193.4 ENST00000375458.2 |
COBLL1 |
cordon-bleu WH2 repeat protein-like 1 |
| chr15_-_83621435 | 0.18 |
ENST00000450735.2 ENST00000426485.1 ENST00000399166.2 ENST00000304231.8 |
HOMER2 |
homer homolog 2 (Drosophila) |
| chr2_+_64681219 | 0.17 |
ENST00000238875.5 |
LGALSL |
lectin, galactoside-binding-like |
| chr3_-_133614297 | 0.17 |
ENST00000486858.1 ENST00000477759.1 |
RAB6B |
RAB6B, member RAS oncogene family |
| chr17_-_54911250 | 0.17 |
ENST00000575658.1 ENST00000397861.2 |
C17orf67 |
chromosome 17 open reading frame 67 |
| chrX_+_150151824 | 0.17 |
ENST00000455596.1 ENST00000448905.2 |
HMGB3 |
high mobility group box 3 |
| chr20_+_814349 | 0.17 |
ENST00000381941.3 |
FAM110A |
family with sequence similarity 110, member A |
| chr20_-_1373606 | 0.17 |
ENST00000381715.1 ENST00000439640.2 ENST00000381719.3 |
FKBP1A |
FK506 binding protein 1A, 12kDa |
| chrX_-_92928557 | 0.17 |
ENST00000373079.3 ENST00000475430.2 |
NAP1L3 |
nucleosome assembly protein 1-like 3 |
| chr3_-_27763803 | 0.17 |
ENST00000449599.1 |
EOMES |
eomesodermin |
| chr11_+_66824346 | 0.16 |
ENST00000532559.1 |
RHOD |
ras homolog family member D |
| chr19_+_34287174 | 0.16 |
ENST00000587559.1 ENST00000588637.1 |
KCTD15 |
potassium channel tetramerization domain containing 15 |
| chr16_+_67063036 | 0.16 |
ENST00000290858.6 ENST00000564034.1 |
CBFB |
core-binding factor, beta subunit |
| chr12_+_122064673 | 0.16 |
ENST00000537188.1 |
ORAI1 |
ORAI calcium release-activated calcium modulator 1 |
| chr17_-_40428359 | 0.16 |
ENST00000293328.3 |
STAT5B |
signal transducer and activator of transcription 5B |
| chr19_+_4343524 | 0.16 |
ENST00000262966.8 ENST00000359935.4 ENST00000599840.1 |
MPND |
MPN domain containing |
| chr15_-_90198659 | 0.16 |
ENST00000394412.3 |
KIF7 |
kinesin family member 7 |
| chr11_-_47207390 | 0.16 |
ENST00000539589.1 ENST00000528462.1 |
PACSIN3 |
protein kinase C and casein kinase substrate in neurons 3 |
| chr9_+_99212403 | 0.16 |
ENST00000375251.3 ENST00000375249.4 |
HABP4 |
hyaluronan binding protein 4 |
| chr8_-_144651024 | 0.16 |
ENST00000524906.1 ENST00000532862.1 ENST00000534459.1 |
MROH6 |
maestro heat-like repeat family member 6 |
| chr20_-_30310693 | 0.16 |
ENST00000307677.4 ENST00000420653.1 |
BCL2L1 |
BCL2-like 1 |
| chr16_+_9056563 | 0.16 |
ENST00000564485.1 |
RP11-77H9.8 |
RP11-77H9.8 |
| chr7_+_89841144 | 0.16 |
ENST00000394622.2 ENST00000394632.1 ENST00000426158.1 ENST00000394621.2 ENST00000402625.2 |
STEAP2 |
STEAP family member 2, metalloreductase |
| chr19_-_35992780 | 0.16 |
ENST00000593342.1 ENST00000601650.1 ENST00000408915.2 |
DMKN |
dermokine |
| chr12_-_53298841 | 0.16 |
ENST00000293308.6 |
KRT8 |
keratin 8 |
| chr19_+_49458107 | 0.16 |
ENST00000539787.1 ENST00000345358.7 ENST00000391871.3 ENST00000415969.2 ENST00000354470.3 ENST00000506183.1 ENST00000293288.8 |
BAX |
BCL2-associated X protein |
| chr14_+_55033815 | 0.16 |
ENST00000554335.1 |
SAMD4A |
sterile alpha motif domain containing 4A |
| chr10_-_135171510 | 0.16 |
ENST00000278025.4 ENST00000368552.3 |
FUOM |
fucose mutarotase |
| chr17_-_16256718 | 0.15 |
ENST00000476243.1 ENST00000299736.4 |
CENPV |
centromere protein V |
| chr7_+_89841000 | 0.15 |
ENST00000287908.3 |
STEAP2 |
STEAP family member 2, metalloreductase |
| chr7_+_89841024 | 0.15 |
ENST00000394626.1 |
STEAP2 |
STEAP family member 2, metalloreductase |
| chr7_+_128828713 | 0.15 |
ENST00000249373.3 |
SMO |
smoothened, frizzled family receptor |
| chr8_-_103251274 | 0.15 |
ENST00000251810.3 |
RRM2B |
ribonucleotide reductase M2 B (TP53 inducible) |
| chr11_-_64013288 | 0.15 |
ENST00000542235.1 |
PPP1R14B |
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr9_+_132934835 | 0.15 |
ENST00000372398.3 |
NCS1 |
neuronal calcium sensor 1 |
| chr3_-_129035120 | 0.15 |
ENST00000333762.4 |
H1FX |
H1 histone family, member X |
| chr11_+_63580855 | 0.15 |
ENST00000294244.4 |
C11orf84 |
chromosome 11 open reading frame 84 |
| chr2_-_203103185 | 0.15 |
ENST00000409205.1 |
SUMO1 |
small ubiquitin-like modifier 1 |
| chr11_-_68519026 | 0.15 |
ENST00000255087.5 |
MTL5 |
metallothionein-like 5, testis-specific (tesmin) |
| chr1_+_61542922 | 0.15 |
ENST00000407417.3 |
NFIA |
nuclear factor I/A |
| chr20_-_1373726 | 0.15 |
ENST00000400137.4 |
FKBP1A |
FK506 binding protein 1A, 12kDa |
| chr19_-_38743878 | 0.15 |
ENST00000587515.1 |
PPP1R14A |
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chrX_-_37706661 | 0.15 |
ENST00000432389.2 ENST00000378581.3 |
DYNLT3 |
dynein, light chain, Tctex-type 3 |
| chr20_-_35492048 | 0.15 |
ENST00000237536.4 |
SOGA1 |
suppressor of glucose, autophagy associated 1 |
| chr20_+_33464407 | 0.15 |
ENST00000253382.5 |
ACSS2 |
acyl-CoA synthetase short-chain family member 2 |
| chr11_+_65292884 | 0.14 |
ENST00000527009.1 |
SCYL1 |
SCY1-like 1 (S. cerevisiae) |
| chr15_+_41136586 | 0.14 |
ENST00000431806.1 |
SPINT1 |
serine peptidase inhibitor, Kunitz type 1 |
| chr14_+_105886275 | 0.14 |
ENST00000405646.1 |
MTA1 |
metastasis associated 1 |
| chr12_-_113909877 | 0.14 |
ENST00000261731.3 |
LHX5 |
LIM homeobox 5 |
| chr3_+_52017454 | 0.14 |
ENST00000476854.1 ENST00000476351.1 ENST00000494103.1 ENST00000404366.2 ENST00000469863.1 |
ACY1 |
aminoacylase 1 |
| chr17_-_27916621 | 0.14 |
ENST00000225394.3 |
GIT1 |
G protein-coupled receptor kinase interacting ArfGAP 1 |
| chr5_+_139781445 | 0.14 |
ENST00000532219.1 ENST00000394722.3 |
ANKHD1-EIF4EBP3 ANKHD1 |
ANKHD1-EIF4EBP3 readthrough ankyrin repeat and KH domain containing 1 |
| chr5_+_176853702 | 0.14 |
ENST00000507633.1 ENST00000393576.3 ENST00000355958.5 ENST00000528793.1 ENST00000512684.1 |
GRK6 |
G protein-coupled receptor kinase 6 |
| chr16_+_76311169 | 0.14 |
ENST00000307431.8 ENST00000377504.4 |
CNTNAP4 |
contactin associated protein-like 4 |
| chr3_-_185542761 | 0.14 |
ENST00000457616.2 ENST00000346192.3 |
IGF2BP2 |
insulin-like growth factor 2 mRNA binding protein 2 |
| chr20_+_57226284 | 0.14 |
ENST00000458280.1 ENST00000355957.5 ENST00000361770.5 ENST00000312283.8 ENST00000412911.1 ENST00000359617.4 ENST00000371141.4 |
STX16 |
syntaxin 16 |
| chr11_-_71955210 | 0.14 |
ENST00000298231.5 |
PHOX2A |
paired-like homeobox 2a |
| chr20_+_33464238 | 0.14 |
ENST00000360596.2 |
ACSS2 |
acyl-CoA synthetase short-chain family member 2 |
| chr16_-_70285797 | 0.14 |
ENST00000435634.1 |
EXOSC6 |
exosome component 6 |
| chr1_+_41157421 | 0.14 |
ENST00000372654.1 |
NFYC |
nuclear transcription factor Y, gamma |
| chr1_-_54872059 | 0.14 |
ENST00000371320.3 |
SSBP3 |
single stranded DNA binding protein 3 |
| chr19_+_51226573 | 0.14 |
ENST00000250340.4 |
CLEC11A |
C-type lectin domain family 11, member A |
| chr3_+_49507674 | 0.13 |
ENST00000431960.1 ENST00000452317.1 ENST00000435508.2 ENST00000452060.1 ENST00000428779.1 ENST00000419218.1 ENST00000430636.1 |
DAG1 |
dystroglycan 1 (dystrophin-associated glycoprotein 1) |
| chr8_+_95653373 | 0.13 |
ENST00000358397.5 |
ESRP1 |
epithelial splicing regulatory protein 1 |
| chr8_+_95653427 | 0.13 |
ENST00000454170.2 |
ESRP1 |
epithelial splicing regulatory protein 1 |
| chr17_-_27916555 | 0.13 |
ENST00000394869.3 |
GIT1 |
G protein-coupled receptor kinase interacting ArfGAP 1 |
| chrX_-_49041242 | 0.13 |
ENST00000453382.1 ENST00000540849.1 ENST00000536904.1 ENST00000432913.1 |
PRICKLE3 |
prickle homolog 3 (Drosophila) |
| chr4_-_22517620 | 0.13 |
ENST00000502482.1 ENST00000334304.5 |
GPR125 |
G protein-coupled receptor 125 |
| chr15_-_48937884 | 0.13 |
ENST00000560355.1 |
FBN1 |
fibrillin 1 |
| chr16_+_67063142 | 0.13 |
ENST00000412916.2 |
CBFB |
core-binding factor, beta subunit |
| chr16_+_88872176 | 0.13 |
ENST00000569140.1 |
CDT1 |
chromatin licensing and DNA replication factor 1 |
| chr1_+_41157671 | 0.13 |
ENST00000534399.1 ENST00000372653.1 |
NFYC |
nuclear transcription factor Y, gamma |
| chr3_-_185542817 | 0.13 |
ENST00000382199.2 |
IGF2BP2 |
insulin-like growth factor 2 mRNA binding protein 2 |
| chr8_+_95653302 | 0.13 |
ENST00000423620.2 ENST00000433389.2 |
ESRP1 |
epithelial splicing regulatory protein 1 |
| chr5_+_176561129 | 0.13 |
ENST00000511258.1 ENST00000347982.4 |
NSD1 |
nuclear receptor binding SET domain protein 1 |
| chrX_+_24167746 | 0.13 |
ENST00000428571.1 ENST00000539115.1 |
ZFX |
zinc finger protein, X-linked |
| chr9_+_137218362 | 0.13 |
ENST00000481739.1 |
RXRA |
retinoid X receptor, alpha |
| chr7_+_73588575 | 0.13 |
ENST00000265753.8 |
EIF4H |
eukaryotic translation initiation factor 4H |
| chr10_-_135171178 | 0.13 |
ENST00000368551.1 |
FUOM |
fucose mutarotase |
| chr19_-_49567124 | 0.13 |
ENST00000301411.3 |
NTF4 |
neurotrophin 4 |
| chr5_-_132113036 | 0.13 |
ENST00000378706.1 |
SEPT8 |
septin 8 |
| chr8_-_143858590 | 0.13 |
ENST00000398906.1 ENST00000522929.1 |
LYNX1 |
Ly6/neurotoxin 1 |
| chr7_-_73256838 | 0.12 |
ENST00000297873.4 |
WBSCR27 |
Williams Beuren syndrome chromosome region 27 |
| chr6_-_35888824 | 0.12 |
ENST00000361690.3 ENST00000512445.1 |
SRPK1 |
SRSF protein kinase 1 |
| chr1_-_157015162 | 0.12 |
ENST00000368194.3 |
ARHGEF11 |
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr4_+_4861385 | 0.12 |
ENST00000382723.4 |
MSX1 |
msh homeobox 1 |
| chr22_+_45148432 | 0.12 |
ENST00000389774.2 ENST00000396119.2 ENST00000336963.4 ENST00000356099.6 ENST00000412433.1 |
ARHGAP8 |
Rho GTPase activating protein 8 |
| chr8_+_81397876 | 0.12 |
ENST00000430430.1 |
ZBTB10 |
zinc finger and BTB domain containing 10 |
| chr17_-_41738931 | 0.12 |
ENST00000329168.3 ENST00000549132.1 |
MEOX1 |
mesenchyme homeobox 1 |
| chr19_+_34663551 | 0.12 |
ENST00000586157.1 |
LSM14A |
LSM14A, SCD6 homolog A (S. cerevisiae) |
| chr20_+_11871371 | 0.12 |
ENST00000254977.3 |
BTBD3 |
BTB (POZ) domain containing 3 |
| chr6_+_36097992 | 0.12 |
ENST00000211287.4 |
MAPK13 |
mitogen-activated protein kinase 13 |
| chr6_+_41040678 | 0.12 |
ENST00000341376.6 ENST00000353205.5 |
NFYA |
nuclear transcription factor Y, alpha |
| chr1_+_84543821 | 0.12 |
ENST00000370688.3 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
| chr16_-_88772670 | 0.12 |
ENST00000562544.1 |
RNF166 |
ring finger protein 166 |
| chr20_-_30311703 | 0.12 |
ENST00000450273.1 ENST00000456404.1 ENST00000420488.1 ENST00000439267.1 |
BCL2L1 |
BCL2-like 1 |
| chr8_+_21916680 | 0.12 |
ENST00000358242.3 ENST00000415253.1 |
DMTN |
dematin actin binding protein |
| chr5_-_134369973 | 0.12 |
ENST00000265340.7 |
PITX1 |
paired-like homeodomain 1 |
| chr22_+_43506747 | 0.12 |
ENST00000216115.2 |
BIK |
BCL2-interacting killer (apoptosis-inducing) |
| chr16_-_67694129 | 0.12 |
ENST00000602320.1 |
ACD |
adrenocortical dysplasia homolog (mouse) |
| chr14_-_91282726 | 0.12 |
ENST00000328459.6 ENST00000357056.2 |
TTC7B |
tetratricopeptide repeat domain 7B |
| chr19_-_59070239 | 0.12 |
ENST00000595957.1 ENST00000253023.3 |
UBE2M |
ubiquitin-conjugating enzyme E2M |
| chr8_+_21916710 | 0.12 |
ENST00000523266.1 ENST00000519907.1 |
DMTN |
dematin actin binding protein |
| chr16_-_5147743 | 0.12 |
ENST00000587133.1 ENST00000458008.4 ENST00000427587.4 |
FAM86A |
family with sequence similarity 86, member A |
| chr20_+_1246908 | 0.12 |
ENST00000381873.3 ENST00000381867.1 |
SNPH |
syntaphilin |
| chr10_+_12171721 | 0.12 |
ENST00000379020.4 ENST00000379017.3 |
SEC61A2 |
Sec61 alpha 2 subunit (S. cerevisiae) |
| chr16_+_69345243 | 0.12 |
ENST00000254950.11 |
VPS4A |
vacuolar protein sorting 4 homolog A (S. cerevisiae) |
| chr9_+_138371618 | 0.12 |
ENST00000356818.2 |
PPP1R26 |
protein phosphatase 1, regulatory subunit 26 |
| chr9_-_99775862 | 0.12 |
ENST00000602917.1 ENST00000375223.4 |
HIATL2 |
hippocampus abundant transcript-like 2 |
| chr7_+_98476095 | 0.12 |
ENST00000359863.4 ENST00000355540.3 |
TRRAP |
transformation/transcription domain-associated protein |
| chr15_+_49447947 | 0.12 |
ENST00000327171.3 ENST00000560654.1 |
GALK2 |
galactokinase 2 |
| chr9_-_124976185 | 0.12 |
ENST00000464484.2 |
LHX6 |
LIM homeobox 6 |
| chr16_-_67693846 | 0.12 |
ENST00000602850.1 |
ACD |
adrenocortical dysplasia homolog (mouse) |
| chr20_+_32581452 | 0.11 |
ENST00000375114.3 ENST00000448364.1 |
RALY |
RALY heterogeneous nuclear ribonucleoprotein |
| chr12_+_125549925 | 0.11 |
ENST00000316519.6 |
AACS |
acetoacetyl-CoA synthetase |
| chr6_+_36098262 | 0.11 |
ENST00000373761.6 ENST00000373766.5 |
MAPK13 |
mitogen-activated protein kinase 13 |
| chr14_-_105767598 | 0.11 |
ENST00000548421.1 |
BRF1 |
BRF1, RNA polymerase III transcription initiation factor 90 kDa subunit |
| chr6_+_30614779 | 0.11 |
ENST00000293604.6 ENST00000376473.5 |
C6orf136 |
chromosome 6 open reading frame 136 |
| chr5_-_141704566 | 0.11 |
ENST00000344120.4 ENST00000434127.2 |
SPRY4 |
sprouty homolog 4 (Drosophila) |
| chr14_-_36988882 | 0.11 |
ENST00000498187.2 |
NKX2-1 |
NK2 homeobox 1 |
| chr20_+_11871433 | 0.11 |
ENST00000399006.2 ENST00000405977.1 |
BTBD3 |
BTB (POZ) domain containing 3 |
| chr12_-_120907374 | 0.11 |
ENST00000550458.1 |
SRSF9 |
serine/arginine-rich splicing factor 9 |
| chr1_+_41157361 | 0.11 |
ENST00000427410.2 ENST00000447388.3 ENST00000425457.2 ENST00000453631.1 ENST00000456393.2 |
NFYC |
nuclear transcription factor Y, gamma |
| chr5_-_132112921 | 0.11 |
ENST00000378721.4 ENST00000378701.1 |
SEPT8 |
septin 8 |
| chr10_-_133795416 | 0.11 |
ENST00000540159.1 ENST00000368636.4 |
BNIP3 |
BCL2/adenovirus E1B 19kDa interacting protein 3 |
| chr9_+_33817461 | 0.11 |
ENST00000263228.3 |
UBE2R2 |
ubiquitin-conjugating enzyme E2R 2 |
| chr15_+_63414760 | 0.11 |
ENST00000557972.1 |
LACTB |
lactamase, beta |
| chr5_+_126112794 | 0.11 |
ENST00000261366.5 ENST00000395354.1 |
LMNB1 |
lamin B1 |
| chr5_-_141061759 | 0.11 |
ENST00000508305.1 |
ARAP3 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr3_-_178789220 | 0.11 |
ENST00000414084.1 |
ZMAT3 |
zinc finger, matrin-type 3 |
| chr3_-_125094093 | 0.11 |
ENST00000484491.1 ENST00000492394.1 ENST00000471196.1 ENST00000468369.1 ENST00000544464.1 ENST00000485866.1 ENST00000360647.4 |
ZNF148 |
zinc finger protein 148 |
| chr12_+_93965609 | 0.11 |
ENST00000549887.1 ENST00000551556.1 |
SOCS2 |
suppressor of cytokine signaling 2 |
| chr22_-_50699701 | 0.11 |
ENST00000395780.1 |
MAPK12 |
mitogen-activated protein kinase 12 |
| chr22_+_44420149 | 0.11 |
ENST00000338758.7 |
PARVB |
parvin, beta |
| chr1_+_52608046 | 0.11 |
ENST00000357206.2 ENST00000287727.3 |
ZFYVE9 |
zinc finger, FYVE domain containing 9 |
| chr17_+_37618257 | 0.11 |
ENST00000447079.4 |
CDK12 |
cyclin-dependent kinase 12 |
| chr12_+_118814344 | 0.11 |
ENST00000397564.2 |
SUDS3 |
suppressor of defective silencing 3 homolog (S. cerevisiae) |
| chr14_-_104181771 | 0.11 |
ENST00000554913.1 ENST00000554974.1 ENST00000553361.1 ENST00000555055.1 ENST00000555964.1 ENST00000556682.1 ENST00000445556.1 ENST00000553332.1 ENST00000352127.7 |
XRCC3 |
X-ray repair complementing defective repair in Chinese hamster cells 3 |
| chr8_-_101117847 | 0.11 |
ENST00000523287.1 ENST00000519092.1 |
RGS22 |
regulator of G-protein signaling 22 |
| chr13_-_21099935 | 0.11 |
ENST00000298248.7 ENST00000382812.1 |
CRYL1 |
crystallin, lambda 1 |
| chr6_+_2246023 | 0.11 |
ENST00000530833.1 ENST00000525811.1 ENST00000534441.1 ENST00000533653.1 ENST00000534468.1 |
GMDS-AS1 |
GMDS antisense RNA 1 (head to head) |
| chr9_-_25678856 | 0.11 |
ENST00000358022.3 |
TUSC1 |
tumor suppressor candidate 1 |
| chr20_+_34042962 | 0.11 |
ENST00000446710.1 ENST00000420564.1 |
CEP250 |
centrosomal protein 250kDa |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.1 | 0.5 | GO:1903721 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.1 | 0.9 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.1 | 0.3 | GO:0060381 | regulation of single-stranded telomeric DNA binding(GO:0060380) positive regulation of single-stranded telomeric DNA binding(GO:0060381) |
| 0.1 | 0.3 | GO:0000294 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) |
| 0.1 | 0.3 | GO:1902499 | positive regulation of protein autoubiquitination(GO:1902499) |
| 0.1 | 0.2 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.1 | 0.3 | GO:0019541 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate metabolic process(GO:0019541) propionate biosynthetic process(GO:0019542) |
| 0.1 | 0.2 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.1 | 0.5 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.1 | 0.2 | GO:0019605 | butyrate metabolic process(GO:0019605) |
| 0.1 | 0.2 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.1 | 0.2 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.1 | 0.3 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 0.2 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.1 | 0.5 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.1 | 0.2 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.0 | 0.1 | GO:0072579 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.0 | 0.6 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.0 | 0.3 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 0.2 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.1 | GO:0021558 | trochlear nerve development(GO:0021558) |
| 0.0 | 0.1 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.0 | 0.1 | GO:0045994 | positive regulation of translational initiation by iron(GO:0045994) |
| 0.0 | 0.3 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.0 | 0.3 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.0 | 0.2 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.2 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.0 | 0.1 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 0.2 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.0 | 0.1 | GO:1903772 | regulation of viral budding via host ESCRT complex(GO:1903772) |
| 0.0 | 0.1 | GO:0090298 | negative regulation of mitochondrial DNA replication(GO:0090298) negative regulation of mitochondrial DNA metabolic process(GO:1901859) |
| 0.0 | 0.1 | GO:1903542 | negative regulation of exosomal secretion(GO:1903542) |
| 0.0 | 0.1 | GO:0071140 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.0 | 0.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.1 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.0 | 0.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.2 | GO:0019075 | virus maturation(GO:0019075) |
| 0.0 | 0.1 | GO:0003064 | regulation of heart rate by hormone(GO:0003064) |
| 0.0 | 0.2 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate biosynthetic process(GO:0006532) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.0 | GO:2000765 | regulation of cytoplasmic translation(GO:2000765) negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 1.3 | GO:2000810 | regulation of bicellular tight junction assembly(GO:2000810) |
| 0.0 | 0.1 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.0 | 0.3 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 0.2 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) |
| 0.0 | 0.0 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.1 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 0.1 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.3 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.1 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.2 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.0 | 0.1 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.0 | 0.1 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 0.0 | 0.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.1 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.1 | GO:0021919 | BMP signaling pathway involved in spinal cord dorsal/ventral patterning(GO:0021919) |
| 0.0 | 0.2 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.2 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.2 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.0 | 0.2 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.2 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.1 | GO:0043366 | beta selection(GO:0043366) |
| 0.0 | 0.1 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.1 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.1 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.0 | 0.1 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.0 | 0.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.3 | GO:0014010 | Schwann cell proliferation(GO:0014010) |
| 0.0 | 0.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.1 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.1 | GO:0002276 | basophil activation involved in immune response(GO:0002276) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.1 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.1 | GO:1905075 | occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 0.0 | 0.1 | GO:0071484 | cellular response to light intensity(GO:0071484) |
| 0.0 | 0.0 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) |
| 0.0 | 0.1 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.1 | GO:0042351 | 'de novo' GDP-L-fucose biosynthetic process(GO:0042351) |
| 0.0 | 0.1 | GO:0002316 | follicular B cell differentiation(GO:0002316) |
| 0.0 | 0.1 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.0 | 0.1 | GO:1904387 | cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.0 | 0.1 | GO:1904046 | negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 0.0 | 0.0 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0019640 | glucuronate catabolic process(GO:0006064) glucuronate catabolic process to xylulose 5-phosphate(GO:0019640) xylulose 5-phosphate metabolic process(GO:0051167) xylulose 5-phosphate biosynthetic process(GO:1901159) |
| 0.0 | 0.1 | GO:0021898 | commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.1 | GO:0000430 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 | 0.2 | GO:0010635 | regulation of mitochondrial fusion(GO:0010635) |
| 0.0 | 0.1 | GO:2000687 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.0 | 0.0 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.1 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 | 0.1 | GO:0006423 | cysteinyl-tRNA aminoacylation(GO:0006423) |
| 0.0 | 0.0 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 0.1 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) |
| 0.0 | 0.1 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.1 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.1 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 | 0.1 | GO:0015870 | acetylcholine transport(GO:0015870) |
| 0.0 | 0.1 | GO:1901536 | negative regulation of DNA demethylation(GO:1901536) regulation of genetic imprinting(GO:2000653) |
| 0.0 | 0.1 | GO:1900166 | glial cell-derived neurotrophic factor secretion(GO:0044467) regulation of glial cell-derived neurotrophic factor secretion(GO:1900166) positive regulation of glial cell-derived neurotrophic factor secretion(GO:1900168) |
| 0.0 | 0.4 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.1 | GO:0002554 | serotonin production involved in inflammatory response(GO:0002351) serotonin secretion involved in inflammatory response(GO:0002442) serotonin secretion by platelet(GO:0002554) |
| 0.0 | 0.1 | GO:0043000 | Golgi to plasma membrane CFTR protein transport(GO:0043000) |
| 0.0 | 0.1 | GO:0071316 | cellular response to nicotine(GO:0071316) |
| 0.0 | 0.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.1 | GO:0043634 | polyadenylation-dependent ncRNA catabolic process(GO:0043634) |
| 0.0 | 0.2 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.2 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.2 | GO:0033182 | regulation of histone ubiquitination(GO:0033182) |
| 0.0 | 0.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.1 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.0 | 0.2 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.1 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) regulation of histone H3-K14 acetylation(GO:0071440) |
| 0.0 | 0.3 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.2 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.1 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.1 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.2 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.0 | 0.0 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 | 0.2 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.1 | GO:0046833 | positive regulation of RNA export from nucleus(GO:0046833) |
| 0.0 | 0.0 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.0 | GO:1903233 | regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) |
| 0.0 | 0.0 | GO:1904956 | Wnt signaling pathway involved in somitogenesis(GO:0090244) regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
| 0.0 | 0.0 | GO:0061110 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.1 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.0 | GO:0060152 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 | 0.1 | GO:0010710 | regulation of collagen catabolic process(GO:0010710) |
| 0.0 | 0.0 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.1 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.0 | GO:0061535 | glutamate secretion, neurotransmission(GO:0061535) |
| 0.0 | 0.2 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.0 | GO:0098976 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.1 | GO:0030920 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 | 0.1 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 0.4 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.0 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.1 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.0 | 0.2 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.0 | 0.3 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.1 | GO:0044571 | [2Fe-2S] cluster assembly(GO:0044571) |
| 0.0 | 0.1 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.1 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.0 | 0.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.2 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 | 0.1 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 0.0 | GO:1904170 | regulation of bleb assembly(GO:1904170) positive regulation of bleb assembly(GO:1904172) |
| 0.0 | 0.0 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.1 | GO:0010917 | negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.0 | 0.2 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.0 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.0 | 0.1 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) |
| 0.0 | 0.1 | GO:0052405 | negative regulation by host of symbiont molecular function(GO:0052405) |
| 0.0 | 0.3 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.1 | GO:0036496 | regulation of translational initiation by eIF2 alpha dephosphorylation(GO:0036496) |
| 0.0 | 0.0 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 0.0 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.0 | 0.1 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.1 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.0 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.0 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.0 | 0.3 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 0.1 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 0.1 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.0 | 0.0 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.0 | 0.2 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.1 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.1 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.0 | 0.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.2 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.0 | 0.0 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.1 | GO:0001575 | globoside metabolic process(GO:0001575) |
| 0.0 | 0.1 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.0 | 0.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.0 | GO:0032416 | negative regulation of sodium:proton antiporter activity(GO:0032416) |
| 0.0 | 0.1 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 | 0.3 | GO:2000178 | negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.0 | GO:0061734 | parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) |
| 0.0 | 0.0 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.0 | 0.1 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.1 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.0 | 0.8 | GO:0008088 | axo-dendritic transport(GO:0008088) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.1 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.0 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.0 | 0.0 | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) |
| 0.0 | 0.1 | GO:0036018 | cellular response to erythropoietin(GO:0036018) |
| 0.0 | 0.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.1 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.0 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.0 | 0.2 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.1 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) uroporphyrinogen III metabolic process(GO:0046502) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 | 0.0 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.0 | 0.1 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.0 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.0 | 0.0 | GO:0042851 | L-alanine metabolic process(GO:0042851) L-alanine catabolic process(GO:0042853) |
| 0.0 | 0.1 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.0 | GO:0014004 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.0 | 0.0 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 0.0 | 0.0 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.2 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.0 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.0 | 0.1 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.1 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.1 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.0 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.1 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.0 | 0.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 0.0 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.0 | 0.0 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.1 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.0 | 0.2 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.1 | GO:1904386 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.0 | 0.1 | GO:0009304 | tRNA transcription(GO:0009304) |
| 0.0 | 0.2 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 0.1 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.3 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 0.0 | GO:0090118 | receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 0.0 | 0.0 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 0.1 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.1 | 0.9 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 1.0 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 0.3 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 0.2 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.2 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.0 | 0.1 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.0 | 0.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 0.1 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.2 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.1 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 0.1 | GO:0031379 | RNA-directed RNA polymerase complex(GO:0031379) |
| 0.0 | 0.1 | GO:0016938 | kinesin I complex(GO:0016938) |
| 0.0 | 0.1 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.1 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.1 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.0 | 0.1 | GO:0030849 | autosome(GO:0030849) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.3 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.1 | GO:0000229 | cytoplasmic chromosome(GO:0000229) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.2 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.1 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 0.0 | 0.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.1 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.1 | GO:0034680 | integrin alpha1-beta1 complex(GO:0034665) integrin alpha3-beta1 complex(GO:0034667) integrin alpha10-beta1 complex(GO:0034680) integrin alpha11-beta1 complex(GO:0034681) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.1 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.7 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.3 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.0 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.1 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 0.2 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.0 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.1 | GO:0000805 | X chromosome(GO:0000805) Barr body(GO:0001740) |
| 0.0 | 0.2 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0097196 | Shu complex(GO:0097196) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.0 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 0.2 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.0 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.0 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.3 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.1 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.1 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.3 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.1 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.1 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.0 | 0.1 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.1 | 0.9 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.2 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.1 | 0.3 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.1 | 0.5 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 0.3 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.4 | GO:0042806 | fucose binding(GO:0042806) |
| 0.1 | 0.2 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.1 | 0.3 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.1 | 0.2 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.1 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) oxidoreductase activity, acting on NAD(P)H, heme protein as acceptor(GO:0016653) |
| 0.0 | 0.1 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.7 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.2 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.0 | 0.1 | GO:0001026 | TFIIIB-type transcription factor activity(GO:0001026) |
| 0.0 | 0.1 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.0 | 0.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.2 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.2 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.2 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.1 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.0 | 0.3 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.0 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.1 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.1 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.0 | 0.2 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.0 | 0.1 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.3 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.2 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.2 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.2 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.5 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.2 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.1 | GO:0031177 | S-acetyltransferase activity(GO:0016418) phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.4 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.1 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.1 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.1 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.2 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 0.1 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.1 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.0 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.1 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.2 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.2 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0004817 | cysteine-tRNA ligase activity(GO:0004817) |
| 0.0 | 0.3 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.0 | 0.1 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.1 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.0 | 0.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.2 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.1 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.1 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.0 | 0.1 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0030572 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.0 | 0.1 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.1 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.0 | 0.1 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.4 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.3 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 1.1 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.1 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.0 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.0 | GO:0097158 | pre-mRNA intronic pyrimidine-rich binding(GO:0097158) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0019825 | oxygen transporter activity(GO:0005344) oxygen binding(GO:0019825) |
| 0.0 | 0.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.1 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.1 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.1 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 1.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.1 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.2 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.2 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.0 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.0 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.0 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.0 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.2 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.0 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.0 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.0 | 0.1 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.2 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.4 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.0 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.1 | GO:0004306 | ethanolamine-phosphate cytidylyltransferase activity(GO:0004306) |
| 0.0 | 0.0 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.1 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.1 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.2 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.0 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.0 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.1 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.0 | 0.1 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.0 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.6 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.0 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.3 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.4 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.2 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.5 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.4 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.1 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 1.1 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.7 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.0 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 0.8 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.1 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.6 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.4 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.4 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.1 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.2 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.2 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.0 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.0 | 0.1 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |