Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for ZIC2_GLI1
Z-value: 0.58
Transcription factors associated with ZIC2_GLI1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
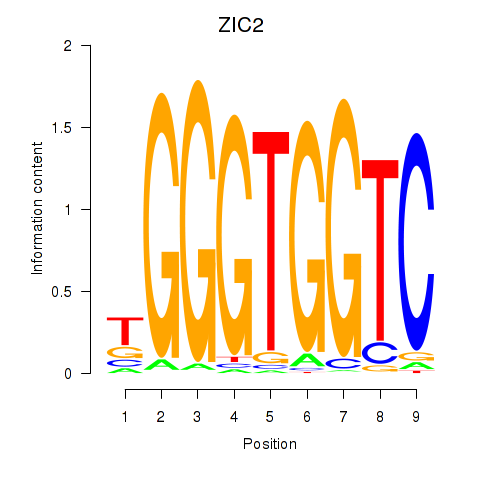
ZIC2
|
ENSG00000043355.6 | ZIC2 |
|
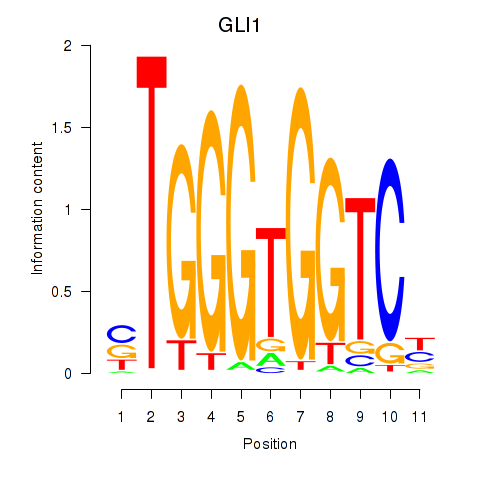
GLI1
|
ENSG00000111087.5 | GLI1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GLI1 | hg19_v2_chr12_+_57857475_57857475 | 0.52 | 1.8e-01 | Click! |
| ZIC2 | hg19_v2_chr13_+_100634004_100634026 | -0.12 | 7.8e-01 | Click! |
Activity profile of ZIC2_GLI1 motif
Sorted Z-values of ZIC2_GLI1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZIC2_GLI1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_51456321 | 0.96 |
ENST00000391809.2 |
KLK5 |
kallikrein-related peptidase 5 |
| chr19_-_51456344 | 0.95 |
ENST00000336334.3 ENST00000593428.1 |
KLK5 |
kallikrein-related peptidase 5 |
| chr9_-_124989804 | 0.52 |
ENST00000373755.2 ENST00000373754.2 |
LHX6 |
LIM homeobox 6 |
| chr8_-_17555164 | 0.45 |
ENST00000297488.6 |
MTUS1 |
microtubule associated tumor suppressor 1 |
| chr5_+_52776449 | 0.42 |
ENST00000396947.3 |
FST |
follistatin |
| chr7_+_69064300 | 0.41 |
ENST00000342771.4 |
AUTS2 |
autism susceptibility candidate 2 |
| chr18_+_21529811 | 0.33 |
ENST00000588004.1 |
LAMA3 |
laminin, alpha 3 |
| chr17_-_34122596 | 0.32 |
ENST00000250144.8 |
MMP28 |
matrix metallopeptidase 28 |
| chr5_+_52776228 | 0.31 |
ENST00000256759.3 |
FST |
follistatin |
| chr8_+_31497271 | 0.31 |
ENST00000520407.1 |
NRG1 |
neuregulin 1 |
| chr11_-_119991589 | 0.30 |
ENST00000526881.1 |
TRIM29 |
tripartite motif containing 29 |
| chr11_-_111783919 | 0.26 |
ENST00000531198.1 ENST00000533879.1 |
CRYAB |
crystallin, alpha B |
| chr7_+_130131907 | 0.24 |
ENST00000223215.4 ENST00000437945.1 |
MEST |
mesoderm specific transcript |
| chr11_-_111783595 | 0.22 |
ENST00000528628.1 |
CRYAB |
crystallin, alpha B |
| chr8_+_75736761 | 0.21 |
ENST00000260113.2 |
PI15 |
peptidase inhibitor 15 |
| chr9_-_98279241 | 0.20 |
ENST00000437951.1 ENST00000375274.2 ENST00000430669.2 ENST00000468211.2 |
PTCH1 |
patched 1 |
| chr11_-_11643540 | 0.20 |
ENST00000227756.4 |
GALNT18 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 18 |
| chr6_+_43739697 | 0.19 |
ENST00000230480.6 |
VEGFA |
vascular endothelial growth factor A |
| chr20_+_44637526 | 0.19 |
ENST00000372330.3 |
MMP9 |
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
| chr2_-_31360887 | 0.18 |
ENST00000420311.2 ENST00000356174.3 ENST00000324589.5 |
GALNT14 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr7_-_108096765 | 0.18 |
ENST00000379024.4 ENST00000351718.4 |
NRCAM |
neuronal cell adhesion molecule |
| chr7_-_108096822 | 0.17 |
ENST00000379028.3 ENST00000413765.2 ENST00000379022.4 |
NRCAM |
neuronal cell adhesion molecule |
| chr1_+_47264711 | 0.17 |
ENST00000371923.4 ENST00000271153.4 ENST00000371919.4 |
CYP4B1 |
cytochrome P450, family 4, subfamily B, polypeptide 1 |
| chr1_+_183155373 | 0.16 |
ENST00000493293.1 ENST00000264144.4 |
LAMC2 |
laminin, gamma 2 |
| chr1_-_12677714 | 0.15 |
ENST00000376223.2 |
DHRS3 |
dehydrogenase/reductase (SDR family) member 3 |
| chr6_+_125540951 | 0.14 |
ENST00000524679.1 |
TPD52L1 |
tumor protein D52-like 1 |
| chr17_-_27278445 | 0.14 |
ENST00000268756.3 ENST00000584685.1 |
PHF12 |
PHD finger protein 12 |
| chr11_+_45944190 | 0.13 |
ENST00000401752.1 ENST00000389968.3 ENST00000325468.5 ENST00000536139.1 |
GYLTL1B |
glycosyltransferase-like 1B |
| chr5_+_161494770 | 0.13 |
ENST00000414552.2 ENST00000361925.4 |
GABRG2 |
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr2_+_106361333 | 0.13 |
ENST00000233154.4 ENST00000451463.2 |
NCK2 |
NCK adaptor protein 2 |
| chr4_-_186697044 | 0.12 |
ENST00000437304.2 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr2_+_96991935 | 0.12 |
ENST00000361124.4 ENST00000420728.1 ENST00000542887.1 |
ITPRIPL1 |
inositol 1,4,5-trisphosphate receptor interacting protein-like 1 |
| chr19_+_45312347 | 0.12 |
ENST00000270233.6 ENST00000591520.1 |
BCAM |
basal cell adhesion molecule (Lutheran blood group) |
| chr20_+_62327996 | 0.12 |
ENST00000369996.1 |
TNFRSF6B |
tumor necrosis factor receptor superfamily, member 6b, decoy |
| chr11_+_111782934 | 0.11 |
ENST00000304298.3 |
HSPB2 |
Homo sapiens heat shock 27kDa protein 2 (HSPB2), mRNA. |
| chr6_+_33172407 | 0.11 |
ENST00000374662.3 |
HSD17B8 |
hydroxysteroid (17-beta) dehydrogenase 8 |
| chrX_+_70364667 | 0.11 |
ENST00000536169.1 ENST00000395855.2 ENST00000374051.3 ENST00000358741.3 |
NLGN3 |
neuroligin 3 |
| chr16_+_2521500 | 0.11 |
ENST00000293973.1 |
NTN3 |
netrin 3 |
| chr10_+_24755416 | 0.11 |
ENST00000396446.1 ENST00000396445.1 ENST00000376451.2 |
KIAA1217 |
KIAA1217 |
| chr4_-_186696425 | 0.11 |
ENST00000430503.1 ENST00000319454.6 ENST00000450341.1 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr6_-_134373732 | 0.10 |
ENST00000275230.5 |
SLC2A12 |
solute carrier family 2 (facilitated glucose transporter), member 12 |
| chr2_+_137523086 | 0.10 |
ENST00000409968.1 |
THSD7B |
thrombospondin, type I, domain containing 7B |
| chr17_+_7211280 | 0.10 |
ENST00000419711.2 ENST00000571955.1 ENST00000573714.1 |
EIF5A |
eukaryotic translation initiation factor 5A |
| chr11_-_6640585 | 0.09 |
ENST00000533371.1 ENST00000528657.1 ENST00000436873.2 ENST00000299427.6 |
TPP1 |
tripeptidyl peptidase I |
| chr2_-_25016251 | 0.09 |
ENST00000328379.5 |
PTRHD1 |
peptidyl-tRNA hydrolase domain containing 1 |
| chr9_+_22446814 | 0.09 |
ENST00000325870.2 |
DMRTA1 |
DMRT-like family A1 |
| chr6_-_32784687 | 0.08 |
ENST00000447394.1 ENST00000438763.2 |
HLA-DOB |
major histocompatibility complex, class II, DO beta |
| chr11_-_111784005 | 0.08 |
ENST00000527899.1 |
CRYAB |
crystallin, alpha B |
| chr22_+_38035459 | 0.08 |
ENST00000357436.4 |
SH3BP1 |
SH3-domain binding protein 1 |
| chr11_-_111782696 | 0.08 |
ENST00000227251.3 ENST00000526180.1 |
CRYAB |
crystallin, alpha B |
| chrX_-_136113833 | 0.08 |
ENST00000298110.1 |
GPR101 |
G protein-coupled receptor 101 |
| chr6_-_19804973 | 0.08 |
ENST00000457670.1 ENST00000607810.1 ENST00000606628.1 |
RP4-625H18.2 |
RP4-625H18.2 |
| chr17_-_37558776 | 0.08 |
ENST00000577399.1 |
FBXL20 |
F-box and leucine-rich repeat protein 20 |
| chr1_-_161014731 | 0.08 |
ENST00000368020.1 |
USF1 |
upstream transcription factor 1 |
| chr20_+_30640004 | 0.07 |
ENST00000520553.1 ENST00000518730.1 ENST00000375852.2 |
HCK |
hemopoietic cell kinase |
| chr6_+_163148973 | 0.07 |
ENST00000366888.2 |
PACRG |
PARK2 co-regulated |
| chr19_-_44285401 | 0.07 |
ENST00000262888.3 |
KCNN4 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr9_-_123476612 | 0.07 |
ENST00000426959.1 |
MEGF9 |
multiple EGF-like-domains 9 |
| chr19_-_55881741 | 0.07 |
ENST00000264563.2 ENST00000590625.1 ENST00000585513.1 |
IL11 |
interleukin 11 |
| chr17_+_43239231 | 0.07 |
ENST00000591576.1 ENST00000591070.1 ENST00000592695.1 |
HEXIM2 |
hexamethylene bis-acetamide inducible 2 |
| chr19_-_40919271 | 0.07 |
ENST00000291825.7 ENST00000324001.7 |
PRX |
periaxin |
| chr17_-_27278304 | 0.07 |
ENST00000577226.1 |
PHF12 |
PHD finger protein 12 |
| chr16_-_1031259 | 0.07 |
ENST00000563837.1 ENST00000563863.1 ENST00000565069.1 ENST00000570014.1 |
RP11-161M6.2 LMF1 |
RP11-161M6.2 lipase maturation factor 1 |
| chr1_-_40105617 | 0.07 |
ENST00000372852.3 |
HEYL |
hes-related family bHLH transcription factor with YRPW motif-like |
| chr20_+_30639991 | 0.07 |
ENST00000534862.1 ENST00000538448.1 ENST00000375862.2 |
HCK |
hemopoietic cell kinase |
| chr1_+_24120143 | 0.07 |
ENST00000374501.1 |
LYPLA2 |
lysophospholipase II |
| chr11_-_111782484 | 0.07 |
ENST00000533971.1 |
CRYAB |
crystallin, alpha B |
| chr1_+_209941827 | 0.06 |
ENST00000367023.1 |
TRAF3IP3 |
TRAF3 interacting protein 3 |
| chr17_+_7608511 | 0.06 |
ENST00000226091.2 |
EFNB3 |
ephrin-B3 |
| chr17_+_43238438 | 0.06 |
ENST00000593138.1 ENST00000586681.1 |
HEXIM2 |
hexamethylene bis-acetamide inducible 2 |
| chr1_-_205782304 | 0.06 |
ENST00000367137.3 |
SLC41A1 |
solute carrier family 41 (magnesium transporter), member 1 |
| chr3_-_52486841 | 0.06 |
ENST00000496590.1 |
TNNC1 |
troponin C type 1 (slow) |
| chr20_+_32319463 | 0.06 |
ENST00000342427.2 ENST00000375200.1 |
ZNF341 |
zinc finger protein 341 |
| chr19_+_42772659 | 0.06 |
ENST00000572681.2 |
CIC |
capicua transcriptional repressor |
| chr2_-_51259528 | 0.06 |
ENST00000404971.1 |
NRXN1 |
neurexin 1 |
| chr21_-_27945562 | 0.06 |
ENST00000299340.4 ENST00000435845.2 |
CYYR1 |
cysteine/tyrosine-rich 1 |
| chr7_-_4901625 | 0.06 |
ENST00000404991.1 |
PAPOLB |
poly(A) polymerase beta (testis specific) |
| chr14_-_51561784 | 0.05 |
ENST00000360392.4 |
TRIM9 |
tripartite motif containing 9 |
| chr2_-_85829496 | 0.05 |
ENST00000409668.1 |
TMEM150A |
transmembrane protein 150A |
| chr17_+_43239191 | 0.05 |
ENST00000589230.1 |
HEXIM2 |
hexamethylene bis-acetamide inducible 2 |
| chr17_-_37557846 | 0.05 |
ENST00000394294.3 ENST00000583610.1 ENST00000264658.6 |
FBXL20 |
F-box and leucine-rich repeat protein 20 |
| chr14_+_92980111 | 0.05 |
ENST00000216487.7 ENST00000557762.1 |
RIN3 |
Ras and Rab interactor 3 |
| chr10_-_97050777 | 0.05 |
ENST00000329399.6 |
PDLIM1 |
PDZ and LIM domain 1 |
| chr3_+_6902794 | 0.05 |
ENST00000357716.4 ENST00000486284.1 ENST00000389336.4 ENST00000403881.1 ENST00000402647.2 |
GRM7 |
glutamate receptor, metabotropic 7 |
| chr13_+_20532848 | 0.05 |
ENST00000382874.2 |
ZMYM2 |
zinc finger, MYM-type 2 |
| chr2_-_51259641 | 0.05 |
ENST00000406316.2 ENST00000405581.1 |
NRXN1 |
neurexin 1 |
| chr18_-_12884259 | 0.05 |
ENST00000353319.4 ENST00000327283.3 |
PTPN2 |
protein tyrosine phosphatase, non-receptor type 2 |
| chr1_+_209941942 | 0.05 |
ENST00000487271.1 ENST00000477431.1 |
TRAF3IP3 |
TRAF3 interacting protein 3 |
| chr16_+_30034655 | 0.05 |
ENST00000300575.2 |
C16orf92 |
chromosome 16 open reading frame 92 |
| chr7_-_105516923 | 0.05 |
ENST00000478915.1 |
ATXN7L1 |
ataxin 7-like 1 |
| chr15_+_60296421 | 0.05 |
ENST00000396057.4 |
FOXB1 |
forkhead box B1 |
| chr17_+_4736627 | 0.04 |
ENST00000355280.6 ENST00000347992.7 |
MINK1 |
misshapen-like kinase 1 |
| chr16_-_16317321 | 0.04 |
ENST00000205557.7 ENST00000575728.1 |
ABCC6 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 6 |
| chr10_+_18948311 | 0.04 |
ENST00000377275.3 |
ARL5B |
ADP-ribosylation factor-like 5B |
| chr2_+_159825143 | 0.04 |
ENST00000454300.1 ENST00000263635.6 |
TANC1 |
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chr3_+_4535025 | 0.04 |
ENST00000302640.8 ENST00000354582.6 ENST00000423119.2 ENST00000357086.4 ENST00000456211.2 |
ITPR1 |
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr3_+_170136642 | 0.04 |
ENST00000064724.3 ENST00000486975.1 |
CLDN11 |
claudin 11 |
| chr12_-_57522813 | 0.04 |
ENST00000556155.1 |
STAT6 |
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr10_-_77161650 | 0.04 |
ENST00000372524.4 |
ZNF503 |
zinc finger protein 503 |
| chr4_+_62067860 | 0.04 |
ENST00000514591.1 |
LPHN3 |
latrophilin 3 |
| chr4_-_38666430 | 0.04 |
ENST00000436901.1 |
AC021860.1 |
Uncharacterized protein |
| chr2_-_128643496 | 0.04 |
ENST00000272647.5 |
AMMECR1L |
AMMECR1-like |
| chr20_+_43343886 | 0.04 |
ENST00000190983.4 |
WISP2 |
WNT1 inducible signaling pathway protein 2 |
| chr2_+_68694678 | 0.04 |
ENST00000303795.4 |
APLF |
aprataxin and PNKP like factor |
| chr2_-_51259292 | 0.04 |
ENST00000401669.2 |
NRXN1 |
neurexin 1 |
| chr2_-_51259229 | 0.04 |
ENST00000405472.3 |
NRXN1 |
neurexin 1 |
| chr13_+_20532900 | 0.04 |
ENST00000382871.2 |
ZMYM2 |
zinc finger, MYM-type 2 |
| chr2_+_26915584 | 0.04 |
ENST00000302909.3 |
KCNK3 |
potassium channel, subfamily K, member 3 |
| chr13_+_111767582 | 0.04 |
ENST00000375741.2 ENST00000375739.2 |
ARHGEF7 |
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr17_-_74533963 | 0.04 |
ENST00000293230.5 |
CYGB |
cytoglobin |
| chr11_+_66059339 | 0.04 |
ENST00000327259.4 |
TMEM151A |
transmembrane protein 151A |
| chr1_-_112531777 | 0.04 |
ENST00000315987.2 ENST00000302127.4 |
KCND3 |
potassium voltage-gated channel, Shal-related subfamily, member 3 |
| chr14_-_23834411 | 0.04 |
ENST00000429593.2 |
EFS |
embryonal Fyn-associated substrate |
| chr19_-_14629224 | 0.04 |
ENST00000254322.2 |
DNAJB1 |
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr9_-_34397800 | 0.04 |
ENST00000297623.2 |
C9orf24 |
chromosome 9 open reading frame 24 |
| chr3_+_113616317 | 0.03 |
ENST00000440446.2 ENST00000488680.1 |
GRAMD1C |
GRAM domain containing 1C |
| chr9_-_123476719 | 0.03 |
ENST00000373930.3 |
MEGF9 |
multiple EGF-like-domains 9 |
| chr2_-_208030647 | 0.03 |
ENST00000309446.6 |
KLF7 |
Kruppel-like factor 7 (ubiquitous) |
| chr5_+_176237478 | 0.03 |
ENST00000329542.4 |
UNC5A |
unc-5 homolog A (C. elegans) |
| chr7_+_149412094 | 0.03 |
ENST00000255992.10 ENST00000319551.8 |
KRBA1 |
KRAB-A domain containing 1 |
| chr17_+_4843654 | 0.03 |
ENST00000575111.1 |
RNF167 |
ring finger protein 167 |
| chr20_+_36149602 | 0.03 |
ENST00000062104.2 ENST00000346199.2 |
NNAT |
neuronatin |
| chr19_+_4969116 | 0.03 |
ENST00000588337.1 ENST00000159111.4 ENST00000381759.4 |
KDM4B |
lysine (K)-specific demethylase 4B |
| chr14_+_24583836 | 0.03 |
ENST00000559115.1 ENST00000558215.1 ENST00000557810.1 ENST00000561375.1 ENST00000446197.3 ENST00000559796.1 ENST00000560713.1 ENST00000560901.1 ENST00000559382.1 |
DCAF11 |
DDB1 and CUL4 associated factor 11 |
| chr17_+_4843679 | 0.03 |
ENST00000576229.1 |
RNF167 |
ring finger protein 167 |
| chr16_+_19183671 | 0.03 |
ENST00000562711.2 |
SYT17 |
synaptotagmin XVII |
| chr10_-_95360983 | 0.03 |
ENST00000371464.3 |
RBP4 |
retinol binding protein 4, plasma |
| chr18_-_3880051 | 0.03 |
ENST00000584874.1 |
DLGAP1 |
discs, large (Drosophila) homolog-associated protein 1 |
| chr7_+_119913688 | 0.03 |
ENST00000331113.4 |
KCND2 |
potassium voltage-gated channel, Shal-related subfamily, member 2 |
| chr18_-_31803435 | 0.03 |
ENST00000589544.1 ENST00000269185.4 ENST00000261592.5 |
NOL4 |
nucleolar protein 4 |
| chr20_-_50722183 | 0.03 |
ENST00000371523.4 |
ZFP64 |
ZFP64 zinc finger protein |
| chr12_-_6798523 | 0.03 |
ENST00000319770.3 |
ZNF384 |
zinc finger protein 384 |
| chr12_-_31477072 | 0.03 |
ENST00000454658.2 |
FAM60A |
family with sequence similarity 60, member A |
| chr10_-_69455873 | 0.03 |
ENST00000433211.2 |
CTNNA3 |
catenin (cadherin-associated protein), alpha 3 |
| chr20_+_43343517 | 0.03 |
ENST00000372865.4 |
WISP2 |
WNT1 inducible signaling pathway protein 2 |
| chr12_-_6798616 | 0.03 |
ENST00000355772.4 ENST00000417772.3 ENST00000396801.3 ENST00000396799.2 |
ZNF384 |
zinc finger protein 384 |
| chr1_-_201081579 | 0.03 |
ENST00000367338.3 ENST00000362061.3 |
CACNA1S |
calcium channel, voltage-dependent, L type, alpha 1S subunit |
| chr17_+_17876127 | 0.03 |
ENST00000582416.1 ENST00000313838.8 ENST00000411504.2 ENST00000581264.1 ENST00000399187.1 ENST00000479684.2 ENST00000584166.1 ENST00000585108.1 ENST00000399182.1 ENST00000579977.1 |
LRRC48 |
leucine rich repeat containing 48 |
| chr11_-_64901978 | 0.03 |
ENST00000294256.8 ENST00000377190.3 |
SYVN1 |
synovial apoptosis inhibitor 1, synoviolin |
| chr16_+_56899114 | 0.03 |
ENST00000566786.1 ENST00000438926.2 ENST00000563236.1 ENST00000262502.5 |
SLC12A3 |
solute carrier family 12 (sodium/chloride transporter), member 3 |
| chr11_-_64901900 | 0.03 |
ENST00000526060.1 ENST00000307289.6 ENST00000528487.1 |
SYVN1 |
synovial apoptosis inhibitor 1, synoviolin |
| chr17_+_39261584 | 0.03 |
ENST00000391415.1 |
KRTAP4-9 |
keratin associated protein 4-9 |
| chr5_-_76935513 | 0.03 |
ENST00000306422.3 |
OTP |
orthopedia homeobox |
| chr20_+_43343476 | 0.03 |
ENST00000372868.2 |
WISP2 |
WNT1 inducible signaling pathway protein 2 |
| chr17_+_73997796 | 0.03 |
ENST00000586261.1 |
CDK3 |
cyclin-dependent kinase 3 |
| chr5_-_150460539 | 0.03 |
ENST00000520931.1 ENST00000520695.1 ENST00000521591.1 ENST00000518977.1 |
TNIP1 |
TNFAIP3 interacting protein 1 |
| chr2_-_46769694 | 0.03 |
ENST00000522587.1 |
ATP6V1E2 |
ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E2 |
| chr19_+_36208877 | 0.03 |
ENST00000420124.1 ENST00000222270.7 ENST00000341701.1 |
KMT2B |
Histone-lysine N-methyltransferase 2B |
| chr4_-_176733897 | 0.02 |
ENST00000393658.2 |
GPM6A |
glycoprotein M6A |
| chr20_+_32782375 | 0.02 |
ENST00000568305.1 |
ASIP |
agouti signaling protein |
| chr16_+_86229728 | 0.02 |
ENST00000601250.1 |
LINC01082 |
long intergenic non-protein coding RNA 1082 |
| chr5_-_89705537 | 0.02 |
ENST00000522864.1 ENST00000522083.1 ENST00000522565.1 ENST00000522842.1 ENST00000283122.3 |
CETN3 |
centrin, EF-hand protein, 3 |
| chr5_-_44388899 | 0.02 |
ENST00000264664.4 |
FGF10 |
fibroblast growth factor 10 |
| chr4_+_134070439 | 0.02 |
ENST00000264360.5 |
PCDH10 |
protocadherin 10 |
| chr13_+_20532807 | 0.02 |
ENST00000382869.3 ENST00000382881.3 |
ZMYM2 |
zinc finger, MYM-type 2 |
| chr19_+_58790314 | 0.02 |
ENST00000196548.5 ENST00000608843.1 |
ZNF8 ZNF8 |
Zinc finger protein 8 zinc finger protein 8 |
| chr2_-_28113965 | 0.02 |
ENST00000302188.3 |
RBKS |
ribokinase |
| chr11_-_19223523 | 0.02 |
ENST00000265968.3 |
CSRP3 |
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr5_+_175792459 | 0.02 |
ENST00000310389.5 |
ARL10 |
ADP-ribosylation factor-like 10 |
| chr10_+_99344071 | 0.02 |
ENST00000370647.4 ENST00000370646.4 |
HOGA1 |
4-hydroxy-2-oxoglutarate aldolase 1 |
| chr12_-_6798410 | 0.02 |
ENST00000361959.3 ENST00000436774.2 ENST00000544482.1 |
ZNF384 |
zinc finger protein 384 |
| chr14_+_96968802 | 0.02 |
ENST00000556619.1 ENST00000392990.2 |
PAPOLA |
poly(A) polymerase alpha |
| chr4_+_72052964 | 0.02 |
ENST00000264485.5 ENST00000425175.1 |
SLC4A4 |
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chr9_-_33264676 | 0.02 |
ENST00000472232.3 ENST00000379704.2 |
BAG1 |
BCL2-associated athanogene |
| chr14_+_24584508 | 0.02 |
ENST00000559354.1 ENST00000560459.1 ENST00000559593.1 ENST00000396941.4 ENST00000396936.1 |
DCAF11 |
DDB1 and CUL4 associated factor 11 |
| chr15_-_93616892 | 0.02 |
ENST00000556658.1 ENST00000538818.1 ENST00000425933.2 |
RGMA |
repulsive guidance molecule family member a |
| chr10_-_133109947 | 0.02 |
ENST00000368642.4 |
TCERG1L |
transcription elongation regulator 1-like |
| chr1_-_38512450 | 0.02 |
ENST00000373012.2 |
POU3F1 |
POU class 3 homeobox 1 |
| chr12_-_43945724 | 0.02 |
ENST00000389420.3 ENST00000553158.1 |
ADAMTS20 |
ADAM metallopeptidase with thrombospondin type 1 motif, 20 |
| chr1_-_211752073 | 0.02 |
ENST00000367001.4 |
SLC30A1 |
solute carrier family 30 (zinc transporter), member 1 |
| chr3_-_128902729 | 0.02 |
ENST00000451728.2 ENST00000446936.2 ENST00000502976.1 ENST00000500450.2 ENST00000441626.2 |
CNBP |
CCHC-type zinc finger, nucleic acid binding protein |
| chr9_-_98079965 | 0.02 |
ENST00000289081.3 |
FANCC |
Fanconi anemia, complementation group C |
| chr6_-_32821599 | 0.02 |
ENST00000354258.4 |
TAP1 |
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr3_+_4535155 | 0.02 |
ENST00000544951.1 |
ITPR1 |
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr8_-_41522719 | 0.02 |
ENST00000335651.6 |
ANK1 |
ankyrin 1, erythrocytic |
| chr15_+_81591757 | 0.02 |
ENST00000558332.1 |
IL16 |
interleukin 16 |
| chr3_-_128902759 | 0.02 |
ENST00000422453.2 ENST00000504813.1 ENST00000512338.1 |
CNBP |
CCHC-type zinc finger, nucleic acid binding protein |
| chr9_+_34989638 | 0.02 |
ENST00000453597.3 ENST00000335998.3 ENST00000312316.5 |
DNAJB5 |
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr3_-_72496035 | 0.02 |
ENST00000477973.2 |
RYBP |
RING1 and YY1 binding protein |
| chr15_-_41408339 | 0.02 |
ENST00000401393.3 |
INO80 |
INO80 complex subunit |
| chr10_-_98346801 | 0.02 |
ENST00000371142.4 |
TM9SF3 |
transmembrane 9 superfamily member 3 |
| chr3_+_141496990 | 0.02 |
ENST00000264952.2 |
GRK7 |
G protein-coupled receptor kinase 7 |
| chr3_-_33759699 | 0.02 |
ENST00000399362.4 ENST00000359576.5 ENST00000307312.7 |
CLASP2 |
cytoplasmic linker associated protein 2 |
| chr3_+_136581096 | 0.02 |
ENST00000476286.1 ENST00000488930.1 |
NCK1 |
NCK adaptor protein 1 |
| chr2_-_208989225 | 0.02 |
ENST00000264376.4 |
CRYGD |
crystallin, gamma D |
| chr6_-_32157947 | 0.02 |
ENST00000375050.4 |
PBX2 |
pre-B-cell leukemia homeobox 2 |
| chr9_-_86571628 | 0.02 |
ENST00000376344.3 |
C9orf64 |
chromosome 9 open reading frame 64 |
| chr5_-_36241900 | 0.01 |
ENST00000381937.4 ENST00000514504.1 |
NADK2 |
NAD kinase 2, mitochondrial |
| chr11_+_66036004 | 0.01 |
ENST00000311481.6 ENST00000527397.1 |
RAB1B |
RAB1B, member RAS oncogene family |
| chr16_-_68002456 | 0.01 |
ENST00000576616.1 ENST00000572037.1 ENST00000338335.3 ENST00000422611.2 ENST00000316341.3 |
SLC12A4 |
solute carrier family 12 (potassium/chloride transporter), member 4 |
| chr17_-_26989136 | 0.01 |
ENST00000247020.4 |
SDF2 |
stromal cell-derived factor 2 |
| chr1_+_226736446 | 0.01 |
ENST00000366788.3 ENST00000366789.4 |
C1orf95 |
chromosome 1 open reading frame 95 |
| chr9_+_33265011 | 0.01 |
ENST00000419016.2 |
CHMP5 |
charged multivesicular body protein 5 |
| chr9_+_136399929 | 0.01 |
ENST00000393060.1 |
ADAMTSL2 |
ADAMTS-like 2 |
| chr19_+_1383890 | 0.01 |
ENST00000539480.1 ENST00000313408.7 ENST00000414651.2 |
NDUFS7 |
NADH dehydrogenase (ubiquinone) Fe-S protein 7, 20kDa (NADH-coenzyme Q reductase) |
| chr1_-_156460391 | 0.01 |
ENST00000360595.3 |
MEF2D |
myocyte enhancer factor 2D |
| chr15_+_40861487 | 0.01 |
ENST00000315616.7 ENST00000559271.1 |
RPUSD2 |
RNA pseudouridylate synthase domain containing 2 |
| chrX_-_107979616 | 0.01 |
ENST00000372129.2 |
IRS4 |
insulin receptor substrate 4 |
| chr8_-_71581377 | 0.01 |
ENST00000276590.4 ENST00000522447.1 |
LACTB2 |
lactamase, beta 2 |
| chr8_+_30244580 | 0.01 |
ENST00000523115.1 ENST00000519647.1 |
RBPMS |
RNA binding protein with multiple splicing |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.9 | GO:0002760 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.1 | 0.4 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.1 | 0.7 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 | 0.3 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.1 | 0.2 | GO:1903572 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.1 | 0.2 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.1 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.0 | 0.3 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.5 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.7 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.0 | 0.2 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.0 | 0.0 | GO:1903970 | negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) |
| 0.0 | 0.2 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.2 | GO:0097107 | postsynaptic density assembly(GO:0097107) gephyrin clustering involved in postsynaptic density assembly(GO:0097116) neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.1 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 0.0 | 0.1 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) positive regulation of synaptic vesicle clustering(GO:2000809) positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.0 | 0.1 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.0 | 0.1 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.0 | 0.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0032972 | diaphragm contraction(GO:0002086) regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.0 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.1 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 | 0.1 | GO:0060179 | male mating behavior(GO:0060179) |
| 0.0 | 0.1 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.2 | GO:0007498 | mesoderm development(GO:0007498) |
| 0.0 | 0.0 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.9 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.1 | 0.2 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.0 | 0.3 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.7 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.2 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.3 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.1 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.0 | 0.2 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.3 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.1 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.3 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.0 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.0 | 0.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.6 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.7 | PID BMP PATHWAY | BMP receptor signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |