Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
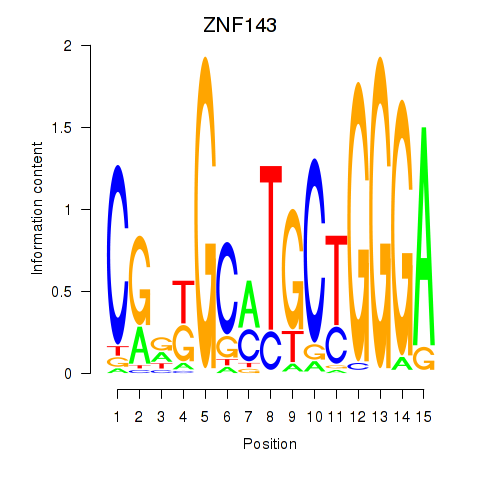
Results for ZNF143
Z-value: 2.12
Transcription factors associated with ZNF143
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF143
|
ENSG00000166478.5 | ZNF143 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF143 | hg19_v2_chr11_+_9482512_9482534 | -0.55 | 1.6e-01 | Click! |
Activity profile of ZNF143 motif
Sorted Z-values of ZNF143 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF143
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_202316392 | 1.27 |
ENST00000194530.3 ENST00000392249.2 |
STRADB |
STE20-related kinase adaptor beta |
| chr2_-_202316260 | 1.14 |
ENST00000332624.3 |
TRAK2 |
trafficking protein, kinesin binding 2 |
| chr15_+_41913690 | 1.13 |
ENST00000563576.1 |
MGA |
MGA, MAX dimerization protein |
| chr19_-_44123734 | 1.07 |
ENST00000598676.1 |
ZNF428 |
zinc finger protein 428 |
| chr19_+_14551066 | 0.98 |
ENST00000342216.4 |
PKN1 |
protein kinase N1 |
| chr17_+_76210367 | 0.98 |
ENST00000592734.1 ENST00000587746.1 |
BIRC5 |
baculoviral IAP repeat containing 5 |
| chr12_+_107168418 | 0.97 |
ENST00000392839.2 ENST00000548914.1 ENST00000355478.2 ENST00000552619.1 ENST00000549643.1 |
RIC8B |
RIC8 guanine nucleotide exchange factor B |
| chr19_-_48673552 | 0.96 |
ENST00000536218.1 ENST00000596549.1 |
LIG1 |
ligase I, DNA, ATP-dependent |
| chr6_-_30709980 | 0.95 |
ENST00000416018.1 ENST00000445853.1 ENST00000413165.1 ENST00000418160.1 |
FLOT1 |
flotillin 1 |
| chr19_+_14544099 | 0.95 |
ENST00000242783.6 ENST00000586557.1 ENST00000590097.1 |
PKN1 |
protein kinase N1 |
| chr10_-_44070016 | 0.91 |
ENST00000374446.2 ENST00000426961.1 ENST00000535642.1 |
ZNF239 |
zinc finger protein 239 |
| chr17_+_76210267 | 0.85 |
ENST00000301633.4 ENST00000350051.3 ENST00000374948.2 ENST00000590449.1 |
BIRC5 |
baculoviral IAP repeat containing 5 |
| chr6_-_30710265 | 0.84 |
ENST00000438162.1 ENST00000454845.1 |
FLOT1 |
flotillin 1 |
| chr19_-_48673580 | 0.84 |
ENST00000427526.2 |
LIG1 |
ligase I, DNA, ATP-dependent |
| chr5_-_150537279 | 0.83 |
ENST00000517486.1 ENST00000377751.5 ENST00000356496.5 ENST00000521512.1 ENST00000517757.1 ENST00000354546.5 |
ANXA6 |
annexin A6 |
| chr2_+_197504278 | 0.81 |
ENST00000272831.7 ENST00000389175.4 ENST00000472405.2 ENST00000423093.2 |
CCDC150 |
coiled-coil domain containing 150 |
| chr12_-_58131931 | 0.78 |
ENST00000547588.1 |
AGAP2 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr20_+_46130671 | 0.77 |
ENST00000371998.3 ENST00000371997.3 |
NCOA3 |
nuclear receptor coactivator 3 |
| chr5_-_64920115 | 0.76 |
ENST00000381018.3 ENST00000274327.7 |
TRIM23 |
tripartite motif containing 23 |
| chr16_+_3074002 | 0.72 |
ENST00000326266.8 ENST00000574549.1 ENST00000575576.1 ENST00000253952.9 |
THOC6 |
THO complex 6 homolog (Drosophila) |
| chr9_-_113800981 | 0.65 |
ENST00000538760.1 |
LPAR1 |
lysophosphatidic acid receptor 1 |
| chr5_-_171433579 | 0.64 |
ENST00000265094.5 ENST00000393802.2 |
FBXW11 |
F-box and WD repeat domain containing 11 |
| chr20_+_46130619 | 0.63 |
ENST00000372004.3 |
NCOA3 |
nuclear receptor coactivator 3 |
| chr15_-_90358048 | 0.63 |
ENST00000300060.6 ENST00000560137.1 |
ANPEP |
alanyl (membrane) aminopeptidase |
| chr6_-_30710510 | 0.63 |
ENST00000376389.3 |
FLOT1 |
flotillin 1 |
| chr20_+_30795664 | 0.63 |
ENST00000375749.3 ENST00000375730.3 ENST00000539210.1 |
POFUT1 |
protein O-fucosyltransferase 1 |
| chr20_+_46130601 | 0.61 |
ENST00000341724.6 |
NCOA3 |
nuclear receptor coactivator 3 |
| chr19_-_37958318 | 0.61 |
ENST00000316950.6 ENST00000591710.1 |
ZNF569 |
zinc finger protein 569 |
| chr12_-_88535747 | 0.61 |
ENST00000309041.7 |
CEP290 |
centrosomal protein 290kDa |
| chr19_-_2456922 | 0.60 |
ENST00000582871.1 ENST00000325327.3 |
LMNB2 |
lamin B2 |
| chr7_-_111846435 | 0.60 |
ENST00000437633.1 ENST00000428084.1 |
DOCK4 |
dedicator of cytokinesis 4 |
| chr19_-_37958086 | 0.60 |
ENST00000592490.1 ENST00000392149.2 |
ZNF569 |
zinc finger protein 569 |
| chr1_-_150693318 | 0.60 |
ENST00000442853.1 ENST00000368995.4 ENST00000368993.2 ENST00000361824.2 ENST00000322343.7 |
HORMAD1 |
HORMA domain containing 1 |
| chr6_-_30710447 | 0.58 |
ENST00000456573.2 |
FLOT1 |
flotillin 1 |
| chr3_+_151986709 | 0.58 |
ENST00000495875.2 ENST00000493459.1 ENST00000324210.5 ENST00000459747.1 |
MBNL1 |
muscleblind-like splicing regulator 1 |
| chr12_+_107168342 | 0.57 |
ENST00000392837.4 |
RIC8B |
RIC8 guanine nucleotide exchange factor B |
| chr2_+_148778570 | 0.55 |
ENST00000407073.1 |
MBD5 |
methyl-CpG binding domain protein 5 |
| chr19_+_37095719 | 0.55 |
ENST00000423582.1 |
ZNF382 |
zinc finger protein 382 |
| chr8_+_27632083 | 0.55 |
ENST00000519637.1 |
ESCO2 |
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr17_-_1619535 | 0.55 |
ENST00000573075.1 ENST00000574306.1 |
MIR22HG |
MIR22 host gene (non-protein coding) |
| chr5_-_171433819 | 0.54 |
ENST00000296933.6 |
FBXW11 |
F-box and WD repeat domain containing 11 |
| chr20_+_30946106 | 0.54 |
ENST00000375687.4 ENST00000542461.1 |
ASXL1 |
additional sex combs like 1 (Drosophila) |
| chr22_-_30162924 | 0.54 |
ENST00000344318.3 ENST00000397781.3 |
ZMAT5 |
zinc finger, matrin-type 5 |
| chr19_+_37096194 | 0.54 |
ENST00000460670.1 ENST00000292928.2 ENST00000439428.1 |
ZNF382 |
zinc finger protein 382 |
| chr8_+_27632047 | 0.54 |
ENST00000397418.2 |
ESCO2 |
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr14_+_104394770 | 0.54 |
ENST00000409874.4 ENST00000339063.5 |
TDRD9 |
tudor domain containing 9 |
| chr9_-_113800317 | 0.54 |
ENST00000374431.3 |
LPAR1 |
lysophosphatidic acid receptor 1 |
| chr5_-_133747551 | 0.53 |
ENST00000395009.3 |
CDKN2AIPNL |
CDKN2A interacting protein N-terminal like |
| chr17_-_1619491 | 0.53 |
ENST00000570416.1 ENST00000575626.1 ENST00000610106.1 ENST00000608198.1 ENST00000609442.1 ENST00000334146.3 ENST00000576489.1 ENST00000608245.1 ENST00000609398.1 ENST00000608913.1 ENST00000574016.1 ENST00000571091.1 ENST00000573127.1 ENST00000609990.1 ENST00000576749.1 |
MIR22HG |
MIR22 host gene (non-protein coding) |
| chr14_-_98444438 | 0.52 |
ENST00000512901.2 |
C14orf64 |
chromosome 14 open reading frame 64 |
| chr12_-_88535842 | 0.51 |
ENST00000550962.1 ENST00000552810.1 |
CEP290 |
centrosomal protein 290kDa |
| chr7_+_65579799 | 0.51 |
ENST00000431089.2 ENST00000398684.2 ENST00000338592.5 |
CRCP |
CGRP receptor component |
| chr1_-_36235529 | 0.49 |
ENST00000318121.3 ENST00000373220.3 ENST00000520551.1 |
CLSPN |
claspin |
| chr1_-_243418344 | 0.49 |
ENST00000366542.1 |
CEP170 |
centrosomal protein 170kDa |
| chr2_+_25015968 | 0.48 |
ENST00000380834.2 ENST00000473706.1 |
CENPO |
centromere protein O |
| chr11_-_82782861 | 0.48 |
ENST00000524635.1 ENST00000526205.1 ENST00000527633.1 ENST00000533486.1 ENST00000533276.2 |
RAB30 |
RAB30, member RAS oncogene family |
| chr15_+_43803143 | 0.48 |
ENST00000382031.1 |
MAP1A |
microtubule-associated protein 1A |
| chr14_+_24025194 | 0.47 |
ENST00000404535.3 ENST00000288014.6 |
THTPA |
thiamine triphosphatase |
| chr21_-_38639601 | 0.47 |
ENST00000539844.1 ENST00000476950.1 ENST00000399001.1 |
DSCR3 |
Down syndrome critical region gene 3 |
| chr12_+_27677085 | 0.46 |
ENST00000545334.1 ENST00000540114.1 ENST00000537927.1 ENST00000318304.8 ENST00000535047.1 ENST00000542629.1 ENST00000228425.6 |
PPFIBP1 |
PTPRF interacting protein, binding protein 1 (liprin beta 1) |
| chr8_-_67525473 | 0.46 |
ENST00000522677.3 |
MYBL1 |
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
| chr12_-_124457257 | 0.45 |
ENST00000545891.1 |
CCDC92 |
coiled-coil domain containing 92 |
| chr16_-_69419473 | 0.45 |
ENST00000566750.1 |
TERF2 |
telomeric repeat binding factor 2 |
| chr3_+_49977440 | 0.45 |
ENST00000442092.1 ENST00000266022.4 ENST00000443081.1 |
RBM6 |
RNA binding motif protein 6 |
| chr19_+_56915668 | 0.45 |
ENST00000333201.9 ENST00000391778.3 |
ZNF583 |
zinc finger protein 583 |
| chr20_+_62289640 | 0.45 |
ENST00000508582.2 ENST00000360203.5 ENST00000356810.4 |
RTEL1 |
regulator of telomere elongation helicase 1 |
| chr15_-_55700216 | 0.43 |
ENST00000569205.1 |
CCPG1 |
cell cycle progression 1 |
| chr12_-_124457371 | 0.43 |
ENST00000238156.3 ENST00000545037.1 |
CCDC92 |
coiled-coil domain containing 92 |
| chr20_-_47804894 | 0.43 |
ENST00000371828.3 ENST00000371856.2 ENST00000360426.4 ENST00000347458.5 ENST00000340954.7 ENST00000371802.1 ENST00000371792.1 ENST00000437404.2 |
STAU1 |
staufen double-stranded RNA binding protein 1 |
| chr12_-_58329819 | 0.42 |
ENST00000551421.1 |
RP11-620J15.3 |
RP11-620J15.3 |
| chr12_+_64798095 | 0.42 |
ENST00000332707.5 |
XPOT |
exportin, tRNA |
| chr6_-_105307711 | 0.42 |
ENST00000519645.1 ENST00000262903.4 ENST00000369125.2 |
HACE1 |
HECT domain and ankyrin repeat containing E3 ubiquitin protein ligase 1 |
| chr11_-_130786333 | 0.42 |
ENST00000533214.1 ENST00000528555.1 ENST00000530356.1 ENST00000539184.1 |
SNX19 |
sorting nexin 19 |
| chr11_-_82782952 | 0.41 |
ENST00000534141.1 |
RAB30 |
RAB30, member RAS oncogene family |
| chr2_-_169746878 | 0.41 |
ENST00000282074.2 |
SPC25 |
SPC25, NDC80 kinetochore complex component |
| chr11_-_113644491 | 0.41 |
ENST00000200135.3 |
ZW10 |
zw10 kinetochore protein |
| chr1_-_243418621 | 0.40 |
ENST00000366544.1 ENST00000366543.1 |
CEP170 |
centrosomal protein 170kDa |
| chr19_+_28284803 | 0.40 |
ENST00000586220.1 ENST00000588784.1 ENST00000591549.1 ENST00000585827.1 ENST00000588636.1 ENST00000587188.1 |
CTC-459F4.3 |
CTC-459F4.3 |
| chr2_-_232645977 | 0.40 |
ENST00000409772.1 |
PDE6D |
phosphodiesterase 6D, cGMP-specific, rod, delta |
| chr12_-_122985494 | 0.39 |
ENST00000336229.4 |
ZCCHC8 |
zinc finger, CCHC domain containing 8 |
| chr9_-_99616642 | 0.39 |
ENST00000478850.1 ENST00000481138.1 |
ZNF782 |
zinc finger protein 782 |
| chr12_+_26111823 | 0.39 |
ENST00000381352.3 ENST00000535907.1 ENST00000405154.2 |
RASSF8 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr2_-_232646015 | 0.39 |
ENST00000287600.4 |
PDE6D |
phosphodiesterase 6D, cGMP-specific, rod, delta |
| chr16_+_2802623 | 0.38 |
ENST00000576924.1 ENST00000575009.1 ENST00000576415.1 ENST00000571378.1 |
SRRM2 |
serine/arginine repetitive matrix 2 |
| chr14_-_89883412 | 0.38 |
ENST00000557258.1 |
FOXN3 |
forkhead box N3 |
| chr12_-_122985067 | 0.38 |
ENST00000540586.1 ENST00000543897.1 |
ZCCHC8 |
zinc finger, CCHC domain containing 8 |
| chr1_+_38259540 | 0.38 |
ENST00000397631.3 |
MANEAL |
mannosidase, endo-alpha-like |
| chr13_-_50367057 | 0.38 |
ENST00000261667.3 |
KPNA3 |
karyopherin alpha 3 (importin alpha 4) |
| chr19_-_38714847 | 0.37 |
ENST00000420980.2 ENST00000355526.4 |
DPF1 |
D4, zinc and double PHD fingers family 1 |
| chr10_+_94352956 | 0.37 |
ENST00000260731.3 |
KIF11 |
kinesin family member 11 |
| chr15_-_101084547 | 0.37 |
ENST00000394113.1 |
CERS3 |
ceramide synthase 3 |
| chr10_+_102672712 | 0.37 |
ENST00000370271.3 ENST00000370269.3 ENST00000609386.1 |
FAM178A |
family with sequence similarity 178, member A |
| chr1_+_38259459 | 0.37 |
ENST00000373045.6 |
MANEAL |
mannosidase, endo-alpha-like |
| chr19_+_54024251 | 0.36 |
ENST00000253144.9 |
ZNF331 |
zinc finger protein 331 |
| chr5_+_64920543 | 0.36 |
ENST00000399438.3 ENST00000510585.2 |
TRAPPC13 CTC-534A2.2 |
trafficking protein particle complex 13 CDNA FLJ26957 fis, clone SLV00486; Uncharacterized protein |
| chr5_-_36152031 | 0.36 |
ENST00000296603.4 |
LMBRD2 |
LMBR1 domain containing 2 |
| chr15_-_101084446 | 0.36 |
ENST00000538112.2 ENST00000559639.1 ENST00000558884.2 |
CERS3 |
ceramide synthase 3 |
| chr8_+_17780483 | 0.35 |
ENST00000517730.1 ENST00000518537.1 ENST00000523055.1 ENST00000519253.1 |
PCM1 |
pericentriolar material 1 |
| chr1_+_45805728 | 0.35 |
ENST00000539779.1 |
TOE1 |
target of EGR1, member 1 (nuclear) |
| chr19_+_37862054 | 0.35 |
ENST00000483919.1 ENST00000588911.1 ENST00000436120.2 ENST00000587349.1 |
ZNF527 |
zinc finger protein 527 |
| chr2_-_27593180 | 0.35 |
ENST00000493344.2 ENST00000445933.2 |
EIF2B4 |
eukaryotic translation initiation factor 2B, subunit 4 delta, 67kDa |
| chr10_+_92631709 | 0.35 |
ENST00000413330.1 ENST00000277882.3 |
RPP30 |
ribonuclease P/MRP 30kDa subunit |
| chr2_+_25016282 | 0.35 |
ENST00000260662.1 |
CENPO |
centromere protein O |
| chr11_+_450255 | 0.35 |
ENST00000308020.5 |
PTDSS2 |
phosphatidylserine synthase 2 |
| chr16_-_25269134 | 0.34 |
ENST00000328086.7 |
ZKSCAN2 |
zinc finger with KRAB and SCAN domains 2 |
| chr8_-_145018905 | 0.34 |
ENST00000398774.2 |
PLEC |
plectin |
| chr12_+_77158021 | 0.34 |
ENST00000550876.1 |
ZDHHC17 |
zinc finger, DHHC-type containing 17 |
| chr9_-_99540328 | 0.34 |
ENST00000223428.4 ENST00000375231.1 ENST00000374641.3 |
ZNF510 |
zinc finger protein 510 |
| chr1_-_19578003 | 0.34 |
ENST00000375199.3 ENST00000375208.3 ENST00000356068.2 ENST00000477853.1 |
EMC1 |
ER membrane protein complex subunit 1 |
| chr4_+_140222609 | 0.34 |
ENST00000296543.5 ENST00000398947.1 |
NAA15 |
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
| chr2_+_172544182 | 0.34 |
ENST00000409197.1 ENST00000456808.1 ENST00000409317.1 ENST00000409773.1 ENST00000411953.1 ENST00000409453.1 |
DYNC1I2 |
dynein, cytoplasmic 1, intermediate chain 2 |
| chr19_-_38270203 | 0.33 |
ENST00000585724.1 ENST00000378445.4 ENST00000588218.1 ENST00000536220.1 ENST00000357309.3 ENST00000339503.4 |
ZNF573 |
zinc finger protein 573 |
| chr2_-_27593306 | 0.33 |
ENST00000347454.4 |
EIF2B4 |
eukaryotic translation initiation factor 2B, subunit 4 delta, 67kDa |
| chr3_+_49977490 | 0.33 |
ENST00000539992.1 |
RBM6 |
RNA binding motif protein 6 |
| chr7_-_134001663 | 0.33 |
ENST00000378509.4 |
SLC35B4 |
solute carrier family 35 (UDP-xylose/UDP-N-acetylglucosamine transporter), member B4 |
| chr3_+_49977523 | 0.33 |
ENST00000422955.1 |
RBM6 |
RNA binding motif protein 6 |
| chr19_-_28284793 | 0.33 |
ENST00000590523.1 |
LINC00662 |
long intergenic non-protein coding RNA 662 |
| chr9_-_127905736 | 0.33 |
ENST00000336505.6 ENST00000373549.4 |
SCAI |
suppressor of cancer cell invasion |
| chr1_+_156052354 | 0.33 |
ENST00000368301.2 |
LMNA |
lamin A/C |
| chr15_-_41099648 | 0.33 |
ENST00000220496.4 |
DNAJC17 |
DnaJ (Hsp40) homolog, subfamily C, member 17 |
| chr12_-_58329888 | 0.33 |
ENST00000546580.1 |
RP11-620J15.3 |
RP11-620J15.3 |
| chr11_+_117198545 | 0.33 |
ENST00000533153.1 ENST00000278935.3 ENST00000525416.1 |
CEP164 |
centrosomal protein 164kDa |
| chr13_+_114145310 | 0.33 |
ENST00000434316.2 ENST00000375391.1 |
TMCO3 |
transmembrane and coiled-coil domains 3 |
| chr22_+_20067738 | 0.32 |
ENST00000351989.3 ENST00000383024.2 |
DGCR8 |
DGCR8 microprocessor complex subunit |
| chr17_+_7590734 | 0.32 |
ENST00000457584.2 |
WRAP53 |
WD repeat containing, antisense to TP53 |
| chr22_-_42342733 | 0.32 |
ENST00000402420.1 |
CENPM |
centromere protein M |
| chr17_+_27369918 | 0.32 |
ENST00000323372.4 |
PIPOX |
pipecolic acid oxidase |
| chr16_-_18937726 | 0.32 |
ENST00000389467.3 ENST00000446231.2 |
SMG1 |
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr1_+_211433275 | 0.32 |
ENST00000367005.4 |
RCOR3 |
REST corepressor 3 |
| chr1_-_35497283 | 0.32 |
ENST00000373333.1 |
ZMYM6 |
zinc finger, MYM-type 6 |
| chr5_-_133747589 | 0.31 |
ENST00000458198.2 |
CDKN2AIPNL |
CDKN2A interacting protein N-terminal like |
| chr2_+_172544011 | 0.31 |
ENST00000508530.1 |
DYNC1I2 |
dynein, cytoplasmic 1, intermediate chain 2 |
| chr5_-_132113559 | 0.31 |
ENST00000448933.1 |
SEPT8 |
septin 8 |
| chr18_+_51884251 | 0.31 |
ENST00000578138.1 |
C18orf54 |
chromosome 18 open reading frame 54 |
| chr19_-_45982076 | 0.31 |
ENST00000423698.2 |
ERCC1 |
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence) |
| chr17_+_38296576 | 0.30 |
ENST00000264645.7 |
CASC3 |
cancer susceptibility candidate 3 |
| chr13_-_107220455 | 0.30 |
ENST00000400198.3 |
ARGLU1 |
arginine and glutamate rich 1 |
| chr19_+_44556158 | 0.30 |
ENST00000434772.3 ENST00000585552.1 |
ZNF223 |
zinc finger protein 223 |
| chrX_-_134049233 | 0.30 |
ENST00000370779.4 |
MOSPD1 |
motile sperm domain containing 1 |
| chr5_+_67584174 | 0.30 |
ENST00000320694.8 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chrX_-_48931648 | 0.29 |
ENST00000376386.3 ENST00000376390.4 |
PRAF2 |
PRA1 domain family, member 2 |
| chr19_-_36909528 | 0.29 |
ENST00000392161.3 ENST00000392171.1 |
ZFP82 |
ZFP82 zinc finger protein |
| chr20_-_35724388 | 0.29 |
ENST00000344359.3 ENST00000373664.3 |
RBL1 |
retinoblastoma-like 1 (p107) |
| chr2_+_172543967 | 0.29 |
ENST00000534253.2 ENST00000263811.4 ENST00000397119.3 ENST00000410079.3 ENST00000438879.1 |
DYNC1I2 |
dynein, cytoplasmic 1, intermediate chain 2 |
| chr15_+_79165222 | 0.29 |
ENST00000559930.1 |
MORF4L1 |
mortality factor 4 like 1 |
| chr12_+_53662110 | 0.29 |
ENST00000552462.1 |
ESPL1 |
extra spindle pole bodies homolog 1 (S. cerevisiae) |
| chr17_-_41277467 | 0.29 |
ENST00000494123.1 ENST00000346315.3 ENST00000309486.4 ENST00000468300.1 ENST00000354071.3 ENST00000352993.3 ENST00000471181.2 |
BRCA1 |
breast cancer 1, early onset |
| chr11_-_65626753 | 0.29 |
ENST00000526975.1 ENST00000531413.1 |
CFL1 |
cofilin 1 (non-muscle) |
| chr9_-_35732362 | 0.28 |
ENST00000314888.9 ENST00000540444.1 |
TLN1 |
talin 1 |
| chr8_-_29120580 | 0.28 |
ENST00000524189.1 |
KIF13B |
kinesin family member 13B |
| chr11_-_790060 | 0.28 |
ENST00000330106.4 |
CEND1 |
cell cycle exit and neuronal differentiation 1 |
| chr2_+_172544294 | 0.28 |
ENST00000358002.6 ENST00000435234.1 ENST00000443458.1 ENST00000412370.1 |
DYNC1I2 |
dynein, cytoplasmic 1, intermediate chain 2 |
| chr2_+_178257372 | 0.28 |
ENST00000264167.4 ENST00000409888.1 |
AGPS |
alkylglycerone phosphate synthase |
| chr5_-_157286104 | 0.28 |
ENST00000530742.1 ENST00000523908.1 ENST00000523094.1 ENST00000296951.5 ENST00000411809.2 |
CLINT1 |
clathrin interactor 1 |
| chr7_+_155437119 | 0.28 |
ENST00000287912.3 |
RBM33 |
RNA binding motif protein 33 |
| chr14_-_65569057 | 0.28 |
ENST00000555419.1 ENST00000341653.2 |
MAX |
MYC associated factor X |
| chr12_+_53662073 | 0.27 |
ENST00000553219.1 ENST00000257934.4 |
ESPL1 |
extra spindle pole bodies homolog 1 (S. cerevisiae) |
| chr17_-_41277370 | 0.27 |
ENST00000476777.1 ENST00000491747.2 ENST00000478531.1 ENST00000477152.1 ENST00000357654.3 ENST00000493795.1 ENST00000493919.1 |
BRCA1 |
breast cancer 1, early onset |
| chr16_+_2802316 | 0.27 |
ENST00000301740.8 |
SRRM2 |
serine/arginine repetitive matrix 2 |
| chr1_+_93646238 | 0.27 |
ENST00000448243.1 ENST00000370276.1 |
CCDC18 |
coiled-coil domain containing 18 |
| chr2_+_172543919 | 0.27 |
ENST00000452242.1 ENST00000340296.4 |
DYNC1I2 |
dynein, cytoplasmic 1, intermediate chain 2 |
| chr2_+_170655789 | 0.27 |
ENST00000409333.1 |
SSB |
Sjogren syndrome antigen B (autoantigen La) |
| chr19_-_38720294 | 0.27 |
ENST00000412732.1 ENST00000456296.1 |
DPF1 |
D4, zinc and double PHD fingers family 1 |
| chr14_+_24025462 | 0.27 |
ENST00000556015.1 ENST00000554970.1 ENST00000554789.1 |
THTPA |
thiamine triphosphatase |
| chr15_-_55790515 | 0.27 |
ENST00000448430.2 ENST00000457155.2 |
DYX1C1 |
dyslexia susceptibility 1 candidate 1 |
| chr5_-_132112921 | 0.27 |
ENST00000378721.4 ENST00000378701.1 |
SEPT8 |
septin 8 |
| chr12_-_56120865 | 0.27 |
ENST00000548898.1 ENST00000552067.1 |
CD63 |
CD63 molecule |
| chr5_-_132113083 | 0.26 |
ENST00000296873.7 |
SEPT8 |
septin 8 |
| chr16_+_67204400 | 0.26 |
ENST00000563439.1 ENST00000432069.2 ENST00000564992.1 ENST00000564053.1 |
NOL3 |
nucleolar protein 3 (apoptosis repressor with CARD domain) |
| chrX_-_119763835 | 0.26 |
ENST00000371313.2 ENST00000304661.5 |
C1GALT1C1 |
C1GALT1-specific chaperone 1 |
| chr19_-_56826157 | 0.26 |
ENST00000592509.1 ENST00000592679.1 ENST00000588442.1 ENST00000593106.1 ENST00000587492.1 ENST00000254165.3 |
ZSCAN5A |
zinc finger and SCAN domain containing 5A |
| chr1_+_28832455 | 0.26 |
ENST00000398958.2 ENST00000427469.1 ENST00000434290.1 ENST00000373833.6 |
RCC1 |
regulator of chromosome condensation 1 |
| chr15_+_40453204 | 0.26 |
ENST00000287598.6 ENST00000412359.3 |
BUB1B |
BUB1 mitotic checkpoint serine/threonine kinase B |
| chr19_-_58609570 | 0.25 |
ENST00000600845.1 ENST00000240727.6 ENST00000600897.1 ENST00000421612.2 ENST00000601063.1 ENST00000601144.1 |
ZSCAN18 |
zinc finger and SCAN domain containing 18 |
| chr1_-_53163992 | 0.25 |
ENST00000371538.3 |
SELRC1 |
cytochrome c oxidase assembly factor 7 |
| chr1_+_32645645 | 0.25 |
ENST00000373609.1 |
TXLNA |
taxilin alpha |
| chr14_-_65569244 | 0.25 |
ENST00000557277.1 ENST00000556892.1 |
MAX |
MYC associated factor X |
| chr13_-_33112823 | 0.25 |
ENST00000504114.1 |
N4BP2L2 |
NEDD4 binding protein 2-like 2 |
| chr19_-_38720354 | 0.24 |
ENST00000416611.1 |
DPF1 |
D4, zinc and double PHD fingers family 1 |
| chr22_+_21336267 | 0.24 |
ENST00000215739.8 |
LZTR1 |
leucine-zipper-like transcription regulator 1 |
| chr17_+_30677136 | 0.24 |
ENST00000394670.4 ENST00000321233.6 ENST00000394673.2 ENST00000341711.6 ENST00000579634.1 ENST00000580759.1 ENST00000342555.6 ENST00000577908.1 ENST00000394679.5 ENST00000582165.1 |
ZNF207 |
zinc finger protein 207 |
| chr11_-_73309228 | 0.24 |
ENST00000356467.4 ENST00000064778.4 |
FAM168A |
family with sequence similarity 168, member A |
| chr16_-_31105870 | 0.24 |
ENST00000394971.3 |
VKORC1 |
vitamin K epoxide reductase complex, subunit 1 |
| chr17_+_48796905 | 0.24 |
ENST00000505658.1 ENST00000393227.2 ENST00000240304.1 ENST00000311571.3 ENST00000505619.1 ENST00000544170.1 ENST00000510984.1 |
LUC7L3 |
LUC7-like 3 (S. cerevisiae) |
| chr19_-_44124019 | 0.24 |
ENST00000300811.3 |
ZNF428 |
zinc finger protein 428 |
| chr2_+_170655322 | 0.23 |
ENST00000260956.4 ENST00000417292.1 |
SSB |
Sjogren syndrome antigen B (autoantigen La) |
| chr1_+_32645269 | 0.23 |
ENST00000373610.3 |
TXLNA |
taxilin alpha |
| chr1_-_213189108 | 0.23 |
ENST00000535388.1 |
ANGEL2 |
angel homolog 2 (Drosophila) |
| chr3_-_9005118 | 0.23 |
ENST00000264926.2 |
RAD18 |
RAD18 homolog (S. cerevisiae) |
| chr17_-_1419878 | 0.23 |
ENST00000449479.1 ENST00000477910.1 ENST00000542125.1 ENST00000575172.1 |
INPP5K |
inositol polyphosphate-5-phosphatase K |
| chr12_+_123011776 | 0.23 |
ENST00000450485.2 ENST00000333479.7 |
KNTC1 |
kinetochore associated 1 |
| chr6_+_56911336 | 0.23 |
ENST00000370733.4 |
KIAA1586 |
KIAA1586 |
| chr11_-_7985193 | 0.23 |
ENST00000328600.2 |
NLRP10 |
NLR family, pyrin domain containing 10 |
| chr5_+_131892603 | 0.23 |
ENST00000378823.3 ENST00000265335.6 |
RAD50 |
RAD50 homolog (S. cerevisiae) |
| chr19_+_47249302 | 0.22 |
ENST00000601299.1 ENST00000318584.5 ENST00000595570.1 ENST00000598271.1 ENST00000597313.1 ENST00000593875.1 ENST00000391909.3 ENST00000602250.1 ENST00000595868.1 ENST00000600629.1 ENST00000602181.1 ENST00000593800.1 ENST00000600227.1 ENST00000600005.1 ENST00000594467.1 ENST00000596460.1 |
FKRP |
fukutin related protein |
| chr12_-_1058849 | 0.22 |
ENST00000358495.3 |
RAD52 |
RAD52 homolog (S. cerevisiae) |
| chr19_-_40791211 | 0.22 |
ENST00000579047.1 |
AKT2 |
v-akt murine thymoma viral oncogene homolog 2 |
| chr1_+_40915725 | 0.22 |
ENST00000484445.1 ENST00000411995.2 ENST00000361584.3 |
ZFP69B |
ZFP69 zinc finger protein B |
| chr16_+_19535235 | 0.22 |
ENST00000565376.2 ENST00000396208.2 |
CCP110 |
centriolar coiled coil protein 110kDa |
| chr18_+_77439775 | 0.22 |
ENST00000299543.7 ENST00000075430.7 |
CTDP1 |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.4 | 1.8 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 0.3 | 3.0 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.3 | 1.9 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.3 | 1.2 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.2 | 1.7 | GO:0035624 | receptor transactivation(GO:0035624) |
| 0.2 | 1.3 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.2 | 0.6 | GO:0051598 | meiotic DNA double-strand break formation(GO:0042138) meiotic recombination checkpoint(GO:0051598) |
| 0.2 | 0.6 | GO:0070512 | regulation of histone H4-K20 methylation(GO:0070510) positive regulation of histone H4-K20 methylation(GO:0070512) |
| 0.2 | 0.7 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.2 | 0.5 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.2 | 0.9 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.2 | 0.6 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.2 | 0.6 | GO:2001151 | regulation of renal water transport(GO:2001151) positive regulation of renal water transport(GO:2001153) |
| 0.1 | 0.6 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.1 | 1.6 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.6 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.5 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.1 | 0.3 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.1 | 0.3 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.1 | 0.4 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.1 | 0.4 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.1 | 0.3 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.1 | 0.6 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.1 | 1.3 | GO:0048793 | pronephros development(GO:0048793) |
| 0.1 | 0.3 | GO:0070631 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.1 | 0.3 | GO:0014876 | response to injury involved in regulation of muscle adaptation(GO:0014876) |
| 0.1 | 1.7 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.1 | 0.6 | GO:2000784 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.1 | 0.4 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 0.7 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.1 | 0.2 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 0.6 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.1 | 0.3 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.1 | 0.2 | GO:0032888 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.1 | 0.3 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.1 | 0.2 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.1 | 0.3 | GO:0019477 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.1 | 0.7 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.1 | 0.2 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 0.1 | 0.4 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 0.1 | 0.2 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.1 | 0.8 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.1 | 0.7 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 0.2 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.1 | 0.3 | GO:0021590 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.1 | 0.3 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.1 | 0.2 | GO:0009644 | response to high light intensity(GO:0009644) cellular response to light intensity(GO:0071484) |
| 0.1 | 0.3 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 0.6 | GO:2000680 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.1 | 0.3 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.1 | 0.9 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 0.2 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.1 | 0.3 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.1 | 0.2 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.1 | 0.2 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.3 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 0.5 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.6 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.2 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.4 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 1.2 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.2 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 0.1 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.0 | 0.3 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.3 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.2 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.1 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.0 | 1.3 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.2 | GO:0001828 | inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 0.0 | 0.1 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.0 | 1.0 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.2 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.3 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.0 | 0.3 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.2 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.1 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.0 | 0.9 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.2 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.3 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 0.1 | GO:1904351 | negative regulation of protein catabolic process in the vacuole(GO:1904351) negative regulation of lysosomal protein catabolic process(GO:1905166) |
| 0.0 | 0.1 | GO:1904382 | protein deglycosylation involved in glycoprotein catabolic process(GO:0035977) glycoprotein ERAD pathway(GO:0097466) mannose trimming involved in glycoprotein ERAD pathway(GO:1904382) |
| 0.0 | 0.5 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.2 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.0 | 0.4 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.1 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.0 | 0.2 | GO:0090656 | t-circle formation(GO:0090656) |
| 0.0 | 0.1 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 1.3 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.2 | GO:0097688 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.0 | 0.1 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.7 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.1 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.2 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.4 | GO:0044818 | mitotic G2 DNA damage checkpoint(GO:0007095) mitotic G2/M transition checkpoint(GO:0044818) |
| 0.0 | 0.1 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.0 | 0.9 | GO:0071173 | mitotic spindle assembly checkpoint(GO:0007094) spindle assembly checkpoint(GO:0071173) |
| 0.0 | 0.3 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.1 | GO:0072554 | blood vessel lumenization(GO:0072554) |
| 0.0 | 0.3 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.1 | GO:0043128 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.0 | 0.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.0 | 0.2 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 0.1 | GO:1903551 | regulation of extracellular exosome assembly(GO:1903551) |
| 0.0 | 0.5 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.2 | GO:2000318 | positive regulation of T-helper 17 type immune response(GO:2000318) |
| 0.0 | 0.4 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.0 | 0.2 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.2 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.3 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 0.2 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.3 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.3 | GO:0019614 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.0 | 0.1 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.1 | GO:2000323 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.1 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.0 | 0.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.2 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.3 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.0 | 0.1 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.1 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.0 | GO:0038188 | cholecystokinin signaling pathway(GO:0038188) |
| 0.0 | 0.1 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.8 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.3 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.2 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.0 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.0 | 0.2 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.1 | GO:0045005 | replication fork processing(GO:0031297) DNA-dependent DNA replication maintenance of fidelity(GO:0045005) |
| 0.0 | 0.3 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.1 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.1 | GO:0043144 | snoRNA processing(GO:0043144) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.0 | GO:0072240 | DCT cell differentiation(GO:0072069) metanephric DCT cell differentiation(GO:0072240) |
| 0.0 | 0.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.0 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.2 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.4 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.2 | GO:0051984 | positive regulation of chromosome segregation(GO:0051984) |
| 0.0 | 0.1 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.3 | GO:0036344 | platelet morphogenesis(GO:0036344) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.0 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.0 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.2 | 3.0 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.2 | 0.6 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.1 | 0.6 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 0.1 | 0.9 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 0.3 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.1 | 0.5 | GO:0071547 | piP-body(GO:0071547) |
| 0.1 | 0.7 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 0.8 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.2 | GO:1990298 | mitotic checkpoint complex(GO:0033597) bub1-bub3 complex(GO:1990298) |
| 0.1 | 1.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.1 | 0.3 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 0.7 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 0.4 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.1 | 0.2 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.1 | 0.2 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.1 | 1.1 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.2 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 0.3 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.1 | 0.2 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.3 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.6 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.2 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.1 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.0 | 0.3 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 1.6 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.3 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.8 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.4 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 1.3 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.7 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 1.7 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.9 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 1.0 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.4 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 2.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.5 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.8 | GO:0005684 | U2-type spliceosomal complex(GO:0005684) |
| 0.0 | 0.1 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.7 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.1 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.0 | 0.2 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.7 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.1 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 0.0 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.1 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.1 | GO:0035061 | perichromatin fibrils(GO:0005726) interchromatin granule(GO:0035061) |
| 0.0 | 1.1 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.8 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 1.2 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 1.0 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.0 | 1.9 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.6 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.3 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 1.5 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 0.0 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.3 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 2.8 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 0.0 | 0.8 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.6 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 1.0 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.5 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.2 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.9 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.2 | 1.8 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.2 | 1.3 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.6 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.7 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.1 | 1.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.3 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.1 | 0.3 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.1 | 0.6 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 0.8 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.1 | 1.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 0.2 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.1 | 0.9 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.2 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.1 | 0.3 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.1 | 1.5 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 0.3 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.1 | 2.0 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 0.2 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.1 | 0.2 | GO:0032408 | MutLbeta complex binding(GO:0032406) MutSbeta complex binding(GO:0032408) |
| 0.1 | 1.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.5 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.1 | 0.3 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.1 | 0.3 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.1 | 0.6 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 2.9 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 0.2 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 0.2 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.0 | 0.5 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.1 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.0 | 0.7 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) |
| 0.0 | 0.2 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.5 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.2 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.3 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.1 | GO:0098626 | methylselenol reductase activity(GO:0098625) methylseleninic acid reductase activity(GO:0098626) |
| 0.0 | 0.4 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 2.1 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.2 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.0 | 0.6 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.3 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.3 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.1 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.0 | 0.5 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.2 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.8 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.2 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.4 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.1 | GO:0000701 | purine-specific mismatch base pair DNA N-glycosylase activity(GO:0000701) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.3 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.2 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.3 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.4 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.1 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.0 | 0.3 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.2 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.3 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.0 | 0.2 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.1 | GO:0003955 | NAD(P)H dehydrogenase (quinone) activity(GO:0003955) 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.0 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.2 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.1 | GO:0004960 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.0 | 0.3 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.6 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0052642 | lysophosphatidic acid phosphatase activity(GO:0052642) |
| 0.0 | 0.2 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.0 | 0.2 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.2 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.7 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.1 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.0 | 0.2 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0004793 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.7 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.2 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.2 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.7 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.1 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.7 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.7 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.0 | GO:0004951 | cholecystokinin receptor activity(GO:0004951) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 1.0 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.1 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.3 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.4 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.2 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.0 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.0 | 0.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.2 | GO:0015245 | fatty acid transporter activity(GO:0015245) very long-chain fatty acid-CoA ligase activity(GO:0031957) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 2.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 0.1 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 3.0 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 3.3 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.4 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 3.0 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.1 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.2 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 1.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.2 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.6 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.4 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.4 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.5 | PID RHOA PATHWAY | RhoA signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 2.0 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 1.3 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.7 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 5.1 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 3.4 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.6 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.7 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.4 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.2 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.0 | 0.3 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.5 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.5 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.5 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.3 | REACTOME TRANSCRIPTION COUPLED NER TC NER | Genes involved in Transcription-coupled NER (TC-NER) |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.5 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.2 | REACTOME RESOLUTION OF AP SITES VIA THE MULTIPLE NUCLEOTIDE PATCH REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the multiple-nucleotide patch replacement pathway |
| 0.0 | 0.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.0 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |