Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
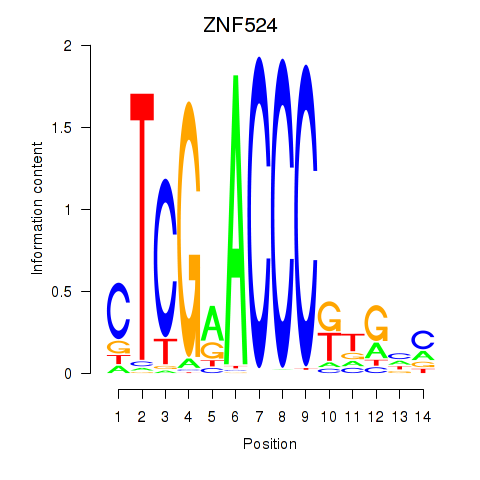
Results for ZNF524
Z-value: 0.54
Transcription factors associated with ZNF524
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF524
|
ENSG00000171443.6 | ZNF524 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF524 | hg19_v2_chr19_+_56111680_56111735 | 0.86 | 5.5e-03 | Click! |
Activity profile of ZNF524 motif
Sorted Z-values of ZNF524 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF524
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_57335280 | 0.52 |
ENST00000287156.4 |
UBE2L6 |
ubiquitin-conjugating enzyme E2L 6 |
| chr19_+_49458107 | 0.44 |
ENST00000539787.1 ENST00000345358.7 ENST00000391871.3 ENST00000415969.2 ENST00000354470.3 ENST00000506183.1 ENST00000293288.8 |
BAX |
BCL2-associated X protein |
| chrX_-_107018969 | 0.39 |
ENST00000372383.4 |
TSC22D3 |
TSC22 domain family, member 3 |
| chr2_+_48757278 | 0.32 |
ENST00000404752.1 ENST00000406226.1 |
STON1 |
stonin 1 |
| chrX_-_107019181 | 0.32 |
ENST00000315660.4 ENST00000372384.2 ENST00000502650.1 ENST00000506724.1 |
TSC22D3 |
TSC22 domain family, member 3 |
| chr11_-_57335750 | 0.30 |
ENST00000340573.4 |
UBE2L6 |
ubiquitin-conjugating enzyme E2L 6 |
| chr20_+_62694590 | 0.30 |
ENST00000339217.4 |
TCEA2 |
transcription elongation factor A (SII), 2 |
| chr19_-_47735918 | 0.28 |
ENST00000449228.1 ENST00000300880.7 ENST00000341983.4 |
BBC3 |
BCL2 binding component 3 |
| chr11_-_65816591 | 0.26 |
ENST00000312006.4 |
GAL3ST3 |
galactose-3-O-sulfotransferase 3 |
| chr20_+_62694461 | 0.25 |
ENST00000343484.5 ENST00000395053.3 |
TCEA2 |
transcription elongation factor A (SII), 2 |
| chr19_+_11457232 | 0.25 |
ENST00000587531.1 |
CCDC159 |
coiled-coil domain containing 159 |
| chr20_+_43160409 | 0.25 |
ENST00000372894.3 ENST00000372892.3 ENST00000372891.3 |
PKIG |
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr2_-_27294500 | 0.24 |
ENST00000447619.1 ENST00000429985.1 ENST00000456793.1 |
OST4 |
oligosaccharyltransferase 4 homolog (S. cerevisiae) |
| chr2_+_64681103 | 0.24 |
ENST00000464281.1 |
LGALSL |
lectin, galactoside-binding-like |
| chr19_-_54984354 | 0.24 |
ENST00000301200.2 |
CDC42EP5 |
CDC42 effector protein (Rho GTPase binding) 5 |
| chr19_+_8478154 | 0.24 |
ENST00000381035.4 ENST00000595142.1 ENST00000601724.1 ENST00000393944.1 ENST00000215555.2 ENST00000601283.1 ENST00000595213.1 |
MARCH2 |
membrane-associated ring finger (C3HC4) 2, E3 ubiquitin protein ligase |
| chr16_+_447209 | 0.24 |
ENST00000382940.4 ENST00000219479.2 |
NME4 |
NME/NM23 nucleoside diphosphate kinase 4 |
| chr19_-_55658650 | 0.23 |
ENST00000589226.1 |
TNNT1 |
troponin T type 1 (skeletal, slow) |
| chr15_+_72410629 | 0.22 |
ENST00000340912.4 ENST00000544171.1 |
SENP8 |
SUMO/sentrin specific peptidase family member 8 |
| chr16_+_31119615 | 0.22 |
ENST00000394950.3 ENST00000287507.3 ENST00000219794.6 ENST00000561755.1 |
BCKDK |
branched chain ketoacid dehydrogenase kinase |
| chr2_+_113342011 | 0.22 |
ENST00000324913.5 |
CHCHD5 |
coiled-coil-helix-coiled-coil-helix domain containing 5 |
| chr14_-_89883412 | 0.22 |
ENST00000557258.1 |
FOXN3 |
forkhead box N3 |
| chr22_-_31503490 | 0.22 |
ENST00000400299.2 |
SELM |
Selenoprotein M |
| chr16_-_4588762 | 0.21 |
ENST00000562334.1 ENST00000562579.1 ENST00000567695.1 ENST00000563507.1 |
CDIP1 |
cell death-inducing p53 target 1 |
| chr17_+_66243715 | 0.21 |
ENST00000359904.3 |
AMZ2 |
archaelysin family metallopeptidase 2 |
| chr19_-_55658281 | 0.21 |
ENST00000585321.2 ENST00000587465.2 |
TNNT1 |
troponin T type 1 (skeletal, slow) |
| chr16_-_4588822 | 0.21 |
ENST00000564828.1 |
CDIP1 |
cell death-inducing p53 target 1 |
| chr17_+_9728828 | 0.20 |
ENST00000262441.5 |
GLP2R |
glucagon-like peptide 2 receptor |
| chr4_-_57522673 | 0.20 |
ENST00000381255.3 ENST00000317745.7 ENST00000555760.2 ENST00000556614.2 |
HOPX |
HOP homeobox |
| chr1_-_3528034 | 0.20 |
ENST00000356575.4 |
MEGF6 |
multiple EGF-like-domains 6 |
| chrX_+_135251835 | 0.20 |
ENST00000456445.1 |
FHL1 |
four and a half LIM domains 1 |
| chr16_+_4743688 | 0.20 |
ENST00000304301.6 ENST00000586252.1 |
NUDT16L1 |
nudix (nucleoside diphosphate linked moiety X)-type motif 16-like 1 |
| chr5_-_174871136 | 0.20 |
ENST00000393752.2 |
DRD1 |
dopamine receptor D1 |
| chr20_+_43160458 | 0.20 |
ENST00000372889.1 ENST00000372887.1 ENST00000372882.3 |
PKIG |
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr2_+_23608064 | 0.19 |
ENST00000486442.1 |
KLHL29 |
kelch-like family member 29 |
| chr1_+_70820451 | 0.19 |
ENST00000361764.4 ENST00000359875.5 ENST00000370940.5 ENST00000531950.1 ENST00000432224.1 |
HHLA3 |
HERV-H LTR-associating 3 |
| chr5_-_111093759 | 0.19 |
ENST00000509979.1 ENST00000513100.1 ENST00000508161.1 ENST00000455559.2 |
NREP |
neuronal regeneration related protein |
| chr12_+_120740119 | 0.19 |
ENST00000536460.1 ENST00000202967.4 |
SIRT4 |
sirtuin 4 |
| chr12_+_7013897 | 0.18 |
ENST00000007969.8 ENST00000323702.5 |
LRRC23 |
leucine rich repeat containing 23 |
| chr19_-_45681482 | 0.18 |
ENST00000592647.1 ENST00000006275.4 ENST00000588062.1 ENST00000585934.1 |
TRAPPC6A |
trafficking protein particle complex 6A |
| chr22_+_37415700 | 0.18 |
ENST00000397129.1 |
MPST |
mercaptopyruvate sulfurtransferase |
| chr16_-_4588469 | 0.18 |
ENST00000588381.1 ENST00000563332.2 |
CDIP1 |
cell death-inducing p53 target 1 |
| chr6_+_32811885 | 0.18 |
ENST00000458296.1 ENST00000413039.1 ENST00000429600.1 ENST00000412095.1 ENST00000415067.1 ENST00000395330.1 |
TAPSAR1 PSMB9 |
TAP1 and PSMB8 antisense RNA 1 proteasome (prosome, macropain) subunit, beta type, 9 |
| chr19_+_50016411 | 0.18 |
ENST00000426395.3 ENST00000600273.1 ENST00000599988.1 |
FCGRT |
Fc fragment of IgG, receptor, transporter, alpha |
| chr22_+_37415728 | 0.18 |
ENST00000404802.3 |
MPST |
mercaptopyruvate sulfurtransferase |
| chr14_+_100437780 | 0.18 |
ENST00000402714.2 |
EVL |
Enah/Vasp-like |
| chr10_+_104178946 | 0.17 |
ENST00000432590.1 |
FBXL15 |
F-box and leucine-rich repeat protein 15 |
| chr17_+_44928946 | 0.17 |
ENST00000290015.2 ENST00000393461.2 |
WNT9B |
wingless-type MMTV integration site family, member 9B |
| chr15_-_63449663 | 0.17 |
ENST00000439025.1 |
RPS27L |
ribosomal protein S27-like |
| chr12_-_54867352 | 0.17 |
ENST00000305879.5 |
GTSF1 |
gametocyte specific factor 1 |
| chr17_+_7942424 | 0.17 |
ENST00000573359.1 |
ALOX15B |
arachidonate 15-lipoxygenase, type B |
| chr22_+_37415676 | 0.17 |
ENST00000401419.3 |
MPST |
mercaptopyruvate sulfurtransferase |
| chr22_+_37415776 | 0.17 |
ENST00000341116.3 ENST00000429360.2 ENST00000404393.1 |
MPST |
mercaptopyruvate sulfurtransferase |
| chr2_+_113342163 | 0.17 |
ENST00000409719.1 |
CHCHD5 |
coiled-coil-helix-coiled-coil-helix domain containing 5 |
| chr5_-_168727713 | 0.17 |
ENST00000404867.3 |
SLIT3 |
slit homolog 3 (Drosophila) |
| chrX_+_135251783 | 0.17 |
ENST00000394153.2 |
FHL1 |
four and a half LIM domains 1 |
| chr1_-_182360918 | 0.16 |
ENST00000339526.4 |
GLUL |
glutamate-ammonia ligase |
| chr5_+_81575281 | 0.16 |
ENST00000380167.4 |
ATP6AP1L |
ATPase, H+ transporting, lysosomal accessory protein 1-like |
| chr8_-_135522425 | 0.16 |
ENST00000521673.1 |
ZFAT |
zinc finger and AT hook domain containing |
| chr6_+_32811861 | 0.16 |
ENST00000453426.1 |
TAPSAR1 |
TAP1 and PSMB8 antisense RNA 1 |
| chr9_-_114361919 | 0.16 |
ENST00000422125.1 |
PTGR1 |
prostaglandin reductase 1 |
| chr1_+_150980989 | 0.16 |
ENST00000368935.1 |
PRUNE |
prune exopolyphosphatase |
| chr11_-_66336060 | 0.16 |
ENST00000310325.5 |
CTSF |
cathepsin F |
| chr17_+_57408994 | 0.16 |
ENST00000312655.4 |
YPEL2 |
yippee-like 2 (Drosophila) |
| chr7_-_44365020 | 0.16 |
ENST00000395747.2 ENST00000347193.4 ENST00000346990.4 ENST00000258682.6 ENST00000353625.4 ENST00000421607.1 ENST00000424197.1 ENST00000502837.2 ENST00000350811.3 ENST00000395749.2 |
CAMK2B |
calcium/calmodulin-dependent protein kinase II beta |
| chr1_+_110210644 | 0.16 |
ENST00000369831.2 ENST00000442650.1 ENST00000369827.3 ENST00000460717.3 ENST00000241337.4 ENST00000467579.3 ENST00000414179.2 ENST00000369829.2 |
GSTM2 |
glutathione S-transferase mu 2 (muscle) |
| chr19_-_49522727 | 0.16 |
ENST00000600007.1 |
CTB-60B18.10 |
CTB-60B18.10 |
| chr2_+_64681219 | 0.16 |
ENST00000238875.5 |
LGALSL |
lectin, galactoside-binding-like |
| chr19_-_55972936 | 0.16 |
ENST00000425675.2 ENST00000589080.1 ENST00000085068.3 |
ISOC2 |
isochorismatase domain containing 2 |
| chr14_+_100531615 | 0.16 |
ENST00000392920.3 |
EVL |
Enah/Vasp-like |
| chr9_+_132096166 | 0.16 |
ENST00000436710.1 |
RP11-65J3.1 |
RP11-65J3.1 |
| chr16_+_3115378 | 0.15 |
ENST00000529550.1 ENST00000551122.1 ENST00000525643.2 ENST00000548807.1 ENST00000528163.2 |
IL32 |
interleukin 32 |
| chr17_-_26879567 | 0.15 |
ENST00000581945.1 ENST00000444148.1 ENST00000301032.4 ENST00000335765.4 |
UNC119 |
unc-119 homolog (C. elegans) |
| chr1_+_171060018 | 0.15 |
ENST00000367755.4 ENST00000392085.2 ENST00000542847.1 ENST00000538429.1 ENST00000479749.1 |
FMO3 |
flavin containing monooxygenase 3 |
| chr16_+_67063855 | 0.15 |
ENST00000563939.2 |
CBFB |
core-binding factor, beta subunit |
| chr16_+_4743707 | 0.15 |
ENST00000586536.1 ENST00000405142.1 ENST00000590460.1 |
NUDT16L1 |
nudix (nucleoside diphosphate linked moiety X)-type motif 16-like 1 |
| chr3_+_45071622 | 0.15 |
ENST00000428034.1 |
CLEC3B |
C-type lectin domain family 3, member B |
| chr2_+_220143989 | 0.15 |
ENST00000336576.5 |
DNAJB2 |
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr19_+_47105309 | 0.15 |
ENST00000599839.1 ENST00000596362.1 |
CALM3 |
calmodulin 3 (phosphorylase kinase, delta) |
| chr19_+_17337007 | 0.15 |
ENST00000215061.4 |
OCEL1 |
occludin/ELL domain containing 1 |
| chr5_-_139682658 | 0.15 |
ENST00000524074.1 ENST00000510217.1 ENST00000261813.4 |
PFDN1 |
prefoldin subunit 1 |
| chr2_+_220110177 | 0.15 |
ENST00000409638.3 ENST00000396738.2 ENST00000409516.3 |
STK16 |
serine/threonine kinase 16 |
| chr12_+_7014064 | 0.15 |
ENST00000443597.2 |
LRRC23 |
leucine rich repeat containing 23 |
| chr3_-_156840776 | 0.15 |
ENST00000471357.1 |
LINC00880 |
long intergenic non-protein coding RNA 880 |
| chr7_+_100209725 | 0.15 |
ENST00000223054.4 |
MOSPD3 |
motile sperm domain containing 3 |
| chr8_-_99129338 | 0.15 |
ENST00000520507.1 |
HRSP12 |
heat-responsive protein 12 |
| chr7_+_65958693 | 0.15 |
ENST00000445681.1 ENST00000452565.1 |
GS1-124K5.4 |
GS1-124K5.4 |
| chr16_+_618837 | 0.14 |
ENST00000409439.2 |
PIGQ |
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr18_+_11981547 | 0.14 |
ENST00000588927.1 |
IMPA2 |
inositol(myo)-1(or 4)-monophosphatase 2 |
| chr19_+_50016610 | 0.14 |
ENST00000596975.1 |
FCGRT |
Fc fragment of IgG, receptor, transporter, alpha |
| chr22_-_19166343 | 0.14 |
ENST00000215882.5 |
SLC25A1 |
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1 |
| chr6_-_32811771 | 0.14 |
ENST00000395339.3 ENST00000374882.3 |
PSMB8 |
proteasome (prosome, macropain) subunit, beta type, 8 |
| chrX_+_35816459 | 0.14 |
ENST00000399988.1 ENST00000399992.1 ENST00000399987.1 ENST00000399989.1 |
MAGEB16 |
melanoma antigen family B, 16 |
| chr15_+_25200108 | 0.14 |
ENST00000577949.1 ENST00000338094.6 ENST00000338327.4 ENST00000579070.1 ENST00000577565.1 |
SNURF SNRPN |
SNRPN upstream reading frame protein small nuclear ribonucleoprotein polypeptide N |
| chr14_+_105212297 | 0.14 |
ENST00000556623.1 ENST00000555674.1 |
ADSSL1 |
adenylosuccinate synthase like 1 |
| chr4_+_37003420 | 0.14 |
ENST00000562049.1 |
RP11-103J17.2 |
RP11-103J17.2 |
| chr16_-_686235 | 0.14 |
ENST00000568773.1 ENST00000565163.1 ENST00000397665.2 ENST00000397666.2 ENST00000301686.8 ENST00000338401.4 ENST00000397664.4 ENST00000568830.1 |
C16orf13 |
chromosome 16 open reading frame 13 |
| chr16_+_447226 | 0.14 |
ENST00000433358.1 |
NME4 |
NME/NM23 nucleoside diphosphate kinase 4 |
| chr12_+_12509990 | 0.14 |
ENST00000542728.1 |
LOH12CR1 |
loss of heterozygosity, 12, chromosomal region 1 |
| chr6_+_24667257 | 0.14 |
ENST00000537591.1 ENST00000230048.4 |
ACOT13 |
acyl-CoA thioesterase 13 |
| chrX_+_134555863 | 0.14 |
ENST00000417443.2 |
LINC00086 |
long intergenic non-protein coding RNA 86 |
| chr10_-_98031265 | 0.14 |
ENST00000224337.5 ENST00000371176.2 |
BLNK |
B-cell linker |
| chr17_-_15587602 | 0.14 |
ENST00000416464.2 ENST00000578237.1 ENST00000581200.1 |
TRIM16 |
tripartite motif containing 16 |
| chr19_+_41092680 | 0.14 |
ENST00000594298.1 ENST00000597396.1 |
SHKBP1 |
SH3KBP1 binding protein 1 |
| chr8_-_99129384 | 0.14 |
ENST00000521560.1 ENST00000254878.3 |
HRSP12 |
heat-responsive protein 12 |
| chr1_-_19229248 | 0.14 |
ENST00000375341.3 |
ALDH4A1 |
aldehyde dehydrogenase 4 family, member A1 |
| chr2_-_232395169 | 0.14 |
ENST00000305141.4 |
NMUR1 |
neuromedin U receptor 1 |
| chr5_-_172198190 | 0.14 |
ENST00000239223.3 |
DUSP1 |
dual specificity phosphatase 1 |
| chr10_-_98031310 | 0.14 |
ENST00000427367.2 ENST00000413476.2 |
BLNK |
B-cell linker |
| chr1_+_26147319 | 0.13 |
ENST00000374300.3 |
MTFR1L |
mitochondrial fission regulator 1-like |
| chr16_-_18801643 | 0.13 |
ENST00000322989.4 ENST00000563390.1 |
RPS15A |
ribosomal protein S15a |
| chr6_+_168841817 | 0.13 |
ENST00000356284.2 ENST00000354536.5 |
SMOC2 |
SPARC related modular calcium binding 2 |
| chr8_-_95961578 | 0.13 |
ENST00000448464.2 ENST00000342697.4 |
TP53INP1 |
tumor protein p53 inducible nuclear protein 1 |
| chr9_-_77703115 | 0.13 |
ENST00000361092.4 ENST00000376808.4 |
NMRK1 |
nicotinamide riboside kinase 1 |
| chr4_+_3388057 | 0.13 |
ENST00000538395.1 |
RGS12 |
regulator of G-protein signaling 12 |
| chr2_+_64681641 | 0.13 |
ENST00000409537.2 |
LGALSL |
lectin, galactoside-binding-like |
| chr17_+_66244071 | 0.13 |
ENST00000580548.1 ENST00000580753.1 ENST00000392720.2 ENST00000359783.4 ENST00000584837.1 ENST00000579724.1 ENST00000584494.1 ENST00000580837.1 |
AMZ2 |
archaelysin family metallopeptidase 2 |
| chr19_-_40931891 | 0.13 |
ENST00000357949.4 |
SERTAD1 |
SERTA domain containing 1 |
| chr19_+_1104415 | 0.13 |
ENST00000585362.2 |
GPX4 |
glutathione peroxidase 4 |
| chr2_+_61372226 | 0.13 |
ENST00000426997.1 |
C2orf74 |
chromosome 2 open reading frame 74 |
| chr7_+_140396946 | 0.13 |
ENST00000476470.1 ENST00000471136.1 |
NDUFB2 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa |
| chr1_+_150980889 | 0.13 |
ENST00000450884.1 ENST00000271620.3 ENST00000271619.8 ENST00000368937.1 ENST00000431193.1 ENST00000368936.1 |
PRUNE |
prune exopolyphosphatase |
| chr1_+_861095 | 0.13 |
ENST00000342066.3 |
SAMD11 |
sterile alpha motif domain containing 11 |
| chr19_+_2819854 | 0.13 |
ENST00000317243.5 |
ZNF554 |
zinc finger protein 554 |
| chr14_+_73704201 | 0.13 |
ENST00000340738.5 ENST00000427855.1 ENST00000381166.3 |
PAPLN |
papilin, proteoglycan-like sulfated glycoprotein |
| chr2_+_75185619 | 0.13 |
ENST00000483063.1 |
POLE4 |
polymerase (DNA-directed), epsilon 4, accessory subunit |
| chr19_+_18043810 | 0.13 |
ENST00000445755.2 |
CCDC124 |
coiled-coil domain containing 124 |
| chr19_+_15904761 | 0.12 |
ENST00000308940.8 |
OR10H5 |
olfactory receptor, family 10, subfamily H, member 5 |
| chr2_+_24346324 | 0.12 |
ENST00000407625.1 ENST00000420135.2 |
FAM228B |
family with sequence similarity 228, member B |
| chr10_-_64576105 | 0.12 |
ENST00000242480.3 ENST00000411732.1 |
EGR2 |
early growth response 2 |
| chr17_+_7942335 | 0.12 |
ENST00000380183.4 ENST00000572022.1 ENST00000380173.2 |
ALOX15B |
arachidonate 15-lipoxygenase, type B |
| chr4_+_142142035 | 0.12 |
ENST00000262990.4 ENST00000512809.1 ENST00000503649.1 ENST00000512738.1 ENST00000421169.2 |
ZNF330 |
zinc finger protein 330 |
| chr10_-_71176623 | 0.12 |
ENST00000373306.4 |
TACR2 |
tachykinin receptor 2 |
| chr9_-_77703056 | 0.12 |
ENST00000376811.1 |
NMRK1 |
nicotinamide riboside kinase 1 |
| chr22_-_30162924 | 0.12 |
ENST00000344318.3 ENST00000397781.3 |
ZMAT5 |
zinc finger, matrin-type 5 |
| chr9_+_90112117 | 0.12 |
ENST00000358077.5 |
DAPK1 |
death-associated protein kinase 1 |
| chr21_+_27543175 | 0.12 |
ENST00000608591.1 ENST00000609365.1 |
AP000230.1 |
AP000230.1 |
| chr17_-_1303462 | 0.12 |
ENST00000573026.1 ENST00000575977.1 ENST00000571732.1 ENST00000264335.8 |
YWHAE |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon |
| chr19_-_49137762 | 0.12 |
ENST00000593500.1 |
DBP |
D site of albumin promoter (albumin D-box) binding protein |
| chr9_+_127539481 | 0.12 |
ENST00000373580.3 |
OLFML2A |
olfactomedin-like 2A |
| chr3_-_75834722 | 0.12 |
ENST00000471541.2 |
ZNF717 |
zinc finger protein 717 |
| chr2_+_71205742 | 0.12 |
ENST00000360589.3 |
ANKRD53 |
ankyrin repeat domain 53 |
| chrY_-_297445 | 0.12 |
ENSTR0000390665.3 |
PPP2R3B |
protein phosphatase 2, regulatory subunit B'', beta |
| chr11_-_1780261 | 0.12 |
ENST00000427721.1 |
RP11-295K3.1 |
RP11-295K3.1 |
| chr20_-_34287220 | 0.12 |
ENST00000306750.3 |
NFS1 |
NFS1 cysteine desulfurase |
| chr22_-_46646576 | 0.12 |
ENST00000314567.3 |
CDPF1 |
cysteine-rich, DPF motif domain containing 1 |
| chr19_+_5823813 | 0.12 |
ENST00000303212.2 |
NRTN |
neurturin |
| chr2_+_55746722 | 0.12 |
ENST00000339012.3 |
CCDC104 |
coiled-coil domain containing 104 |
| chr4_+_62067860 | 0.12 |
ENST00000514591.1 |
LPHN3 |
latrophilin 3 |
| chr12_-_101801505 | 0.12 |
ENST00000539055.1 ENST00000551688.1 ENST00000551671.1 ENST00000261636.8 |
ARL1 |
ADP-ribosylation factor-like 1 |
| chrX_+_153492665 | 0.12 |
ENST00000430419.1 |
OPN1MW2 |
opsin 1 (cone pigments), medium-wave-sensitive 2 |
| chr6_-_109330702 | 0.12 |
ENST00000356644.7 |
SESN1 |
sestrin 1 |
| chr19_+_6372444 | 0.12 |
ENST00000245812.3 |
ALKBH7 |
alkB, alkylation repair homolog 7 (E. coli) |
| chr19_-_55658687 | 0.12 |
ENST00000593046.1 |
TNNT1 |
troponin T type 1 (skeletal, slow) |
| chr11_-_207221 | 0.12 |
ENST00000486280.1 ENST00000332865.6 ENST00000529614.2 ENST00000325147.9 ENST00000410108.1 ENST00000382762.3 |
BET1L |
Bet1 golgi vesicular membrane trafficking protein-like |
| chr2_+_30454390 | 0.12 |
ENST00000395323.3 ENST00000406087.1 ENST00000404397.1 |
LBH |
limb bud and heart development |
| chr22_+_46546494 | 0.12 |
ENST00000396000.2 ENST00000262735.5 ENST00000420804.1 |
PPARA |
peroxisome proliferator-activated receptor alpha |
| chr6_+_127588020 | 0.12 |
ENST00000309649.3 ENST00000610162.1 ENST00000610153.1 ENST00000608991.1 ENST00000480444.1 |
RNF146 |
ring finger protein 146 |
| chr3_-_196439065 | 0.12 |
ENST00000399942.4 ENST00000409690.3 |
CEP19 |
centrosomal protein 19kDa |
| chr12_-_56753858 | 0.12 |
ENST00000314128.4 ENST00000557235.1 ENST00000418572.2 |
STAT2 |
signal transducer and activator of transcription 2, 113kDa |
| chr9_+_77703414 | 0.11 |
ENST00000346234.6 |
OSTF1 |
osteoclast stimulating factor 1 |
| chr22_-_24322019 | 0.11 |
ENST00000350608.3 |
DDT |
D-dopachrome tautomerase |
| chr10_-_104474128 | 0.11 |
ENST00000260746.5 |
ARL3 |
ADP-ribosylation factor-like 3 |
| chr1_-_19229014 | 0.11 |
ENST00000538839.1 ENST00000290597.5 |
ALDH4A1 |
aldehyde dehydrogenase 4 family, member A1 |
| chr12_+_12510045 | 0.11 |
ENST00000314565.4 |
LOH12CR1 |
loss of heterozygosity, 12, chromosomal region 1 |
| chr9_-_140351928 | 0.11 |
ENST00000339554.3 |
NSMF |
NMDA receptor synaptonuclear signaling and neuronal migration factor |
| chr14_-_90085458 | 0.11 |
ENST00000345097.4 ENST00000555855.1 ENST00000555353.1 |
FOXN3 |
forkhead box N3 |
| chr16_+_29973351 | 0.11 |
ENST00000602948.1 ENST00000279396.6 ENST00000575829.2 ENST00000561899.2 |
TMEM219 |
transmembrane protein 219 |
| chr12_+_94542459 | 0.11 |
ENST00000258526.4 |
PLXNC1 |
plexin C1 |
| chr1_-_94586651 | 0.11 |
ENST00000535735.1 ENST00000370225.3 |
ABCA4 |
ATP-binding cassette, sub-family A (ABC1), member 4 |
| chr4_-_6675550 | 0.11 |
ENST00000513179.1 ENST00000515205.1 |
RP11-539L10.3 |
RP11-539L10.3 |
| chr1_-_36916066 | 0.11 |
ENST00000315643.9 |
OSCP1 |
organic solute carrier partner 1 |
| chr11_+_64059464 | 0.11 |
ENST00000394525.2 |
KCNK4 |
potassium channel, subfamily K, member 4 |
| chr17_+_79989500 | 0.11 |
ENST00000306897.4 |
RAC3 |
ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) |
| chr20_+_35169885 | 0.11 |
ENST00000279022.2 ENST00000346786.2 |
MYL9 |
myosin, light chain 9, regulatory |
| chr11_-_18034701 | 0.11 |
ENST00000265965.5 |
SERGEF |
secretion regulating guanine nucleotide exchange factor |
| chr3_-_49466686 | 0.11 |
ENST00000273598.3 ENST00000436744.2 |
NICN1 |
nicolin 1 |
| chr22_+_23046750 | 0.11 |
ENST00000390307.2 |
IGLV3-22 |
immunoglobulin lambda variable 3-22 (gene/pseudogene) |
| chr12_-_12509929 | 0.11 |
ENST00000381800.2 |
LOH12CR2 |
loss of heterozygosity, 12, chromosomal region 2 (non-protein coding) |
| chr7_+_44240520 | 0.11 |
ENST00000496112.1 ENST00000223369.2 |
YKT6 |
YKT6 v-SNARE homolog (S. cerevisiae) |
| chr11_-_63993601 | 0.11 |
ENST00000545812.1 ENST00000394547.3 ENST00000317459.6 |
TRPT1 |
tRNA phosphotransferase 1 |
| chr2_+_11752379 | 0.11 |
ENST00000396123.1 |
GREB1 |
growth regulation by estrogen in breast cancer 1 |
| chr14_+_24605389 | 0.11 |
ENST00000382708.3 ENST00000561435.1 |
PSME1 |
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr17_-_79895154 | 0.11 |
ENST00000405481.4 ENST00000585215.1 ENST00000577624.1 ENST00000403172.4 |
PYCR1 |
pyrroline-5-carboxylate reductase 1 |
| chr1_-_182360498 | 0.11 |
ENST00000417584.2 |
GLUL |
glutamate-ammonia ligase |
| chr7_-_142247606 | 0.11 |
ENST00000390361.3 |
TRBV7-3 |
T cell receptor beta variable 7-3 |
| chr10_+_112836779 | 0.10 |
ENST00000280155.2 |
ADRA2A |
adrenoceptor alpha 2A |
| chr17_-_8027402 | 0.10 |
ENST00000541682.2 ENST00000317814.4 ENST00000577735.1 |
HES7 |
hes family bHLH transcription factor 7 |
| chr9_-_138591341 | 0.10 |
ENST00000298466.5 ENST00000425225.1 |
SOHLH1 |
spermatogenesis and oogenesis specific basic helix-loop-helix 1 |
| chr20_-_31331212 | 0.10 |
ENST00000474815.2 ENST00000446419.2 |
COMMD7 |
COMM domain containing 7 |
| chr14_-_92588246 | 0.10 |
ENST00000329559.3 |
NDUFB1 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 1, 7kDa |
| chr12_+_54402790 | 0.10 |
ENST00000040584.4 |
HOXC8 |
homeobox C8 |
| chr1_+_59762642 | 0.10 |
ENST00000371218.4 ENST00000303721.7 |
FGGY |
FGGY carbohydrate kinase domain containing |
| chrX_-_107334750 | 0.10 |
ENST00000340200.5 ENST00000372296.1 ENST00000372295.1 ENST00000361815.5 |
PSMD10 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 |
| chr2_+_54342574 | 0.10 |
ENST00000303536.4 ENST00000394666.3 |
ACYP2 |
acylphosphatase 2, muscle type |
| chr6_-_26056695 | 0.10 |
ENST00000343677.2 |
HIST1H1C |
histone cluster 1, H1c |
| chr1_+_202976493 | 0.10 |
ENST00000367242.3 |
TMEM183A |
transmembrane protein 183A |
| chr16_+_87425914 | 0.10 |
ENST00000565788.1 |
MAP1LC3B |
microtubule-associated protein 1 light chain 3 beta |
| chr5_-_10761206 | 0.10 |
ENST00000432074.2 ENST00000230895.6 |
DAP |
death-associated protein |
| chr19_-_13260992 | 0.10 |
ENST00000242770.5 ENST00000589083.1 ENST00000587230.1 |
STX10 |
syntaxin 10 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0009440 | cyanate metabolic process(GO:0009439) cyanate catabolic process(GO:0009440) |
| 0.1 | 0.4 | GO:1902339 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) positive regulation of apoptotic DNA fragmentation(GO:1902512) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.1 | 0.3 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.9 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.3 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.1 | 0.7 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 0.3 | GO:0018282 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.1 | 0.3 | GO:0071469 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) cellular response to alkaline pH(GO:0071469) |
| 0.1 | 0.3 | GO:0010638 | positive regulation of organelle organization(GO:0010638) |
| 0.1 | 0.2 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.1 | 0.3 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.1 | 0.6 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.1 | 0.2 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.1 | 0.3 | GO:0097327 | response to antineoplastic agent(GO:0097327) |
| 0.1 | 0.3 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) |
| 0.1 | 0.1 | GO:0002682 | regulation of immune system process(GO:0002682) |
| 0.0 | 0.2 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.0 | 0.3 | GO:2001033 | negative regulation of double-strand break repair via nonhomologous end joining(GO:2001033) |
| 0.0 | 0.2 | GO:0019541 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate metabolic process(GO:0019541) propionate biosynthetic process(GO:0019542) |
| 0.0 | 0.1 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 0.0 | 0.1 | GO:0036446 | myofibroblast differentiation(GO:0036446) regulation of myofibroblast differentiation(GO:1904760) |
| 0.0 | 0.1 | GO:1902824 | positive regulation of late endosome to lysosome transport(GO:1902824) |
| 0.0 | 0.3 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.1 | GO:0035283 | rhombomere 3 development(GO:0021569) rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.1 | GO:1902309 | regulation of heart rate by hormone(GO:0003064) negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.2 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.2 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.4 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.1 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.0 | 0.1 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 0.0 | 0.3 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 | 0.1 | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway(GO:0042058) |
| 0.0 | 0.1 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.0 | 0.6 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.4 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.2 | GO:0019343 | cysteine biosynthetic process via cystathionine(GO:0019343) |
| 0.0 | 0.1 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.0 | 0.2 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.4 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.1 | GO:0009436 | cysteine catabolic process(GO:0009093) glyoxylate catabolic process(GO:0009436) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.0 | 0.2 | GO:0021834 | chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) response to cortisol(GO:0051414) apoptotic process involved in luteolysis(GO:0061364) |
| 0.0 | 0.2 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.1 | GO:0021546 | rhombomere development(GO:0021546) |
| 0.0 | 0.1 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.0 | 0.1 | GO:0035625 | negative regulation of epinephrine secretion(GO:0032811) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.0 | 0.2 | GO:0070458 | cellular detoxification of nitrogen compound(GO:0070458) |
| 0.0 | 0.2 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.2 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.0 | 0.1 | GO:0000472 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.0 | 0.1 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.0 | 0.1 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.5 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.2 | GO:0045415 | negative regulation of interleukin-8 biosynthetic process(GO:0045415) |
| 0.0 | 0.1 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.1 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.0 | 0.1 | GO:0070541 | response to platinum ion(GO:0070541) |
| 0.0 | 0.1 | GO:1904674 | positive regulation of somatic stem cell population maintenance(GO:1904674) |
| 0.0 | 0.1 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.0 | 0.0 | GO:1901983 | regulation of protein acetylation(GO:1901983) |
| 0.0 | 0.1 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 0.1 | GO:0042418 | epinephrine biosynthetic process(GO:0042418) |
| 0.0 | 0.0 | GO:0035552 | oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.0 | 0.1 | GO:0031456 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.0 | 0.1 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.0 | 0.1 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.1 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.0 | 0.1 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.1 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.0 | 0.1 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) |
| 0.0 | 0.0 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:2000373 | regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.1 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.0 | 0.2 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 | 0.1 | GO:0019075 | virus maturation(GO:0019075) |
| 0.0 | 0.1 | GO:0001983 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.0 | 0.1 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.0 | 0.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.0 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) |
| 0.0 | 0.4 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.0 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.0 | 0.0 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.0 | 0.2 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.0 | 0.0 | GO:2000547 | regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.0 | 0.1 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.1 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.1 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.0 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 | 0.0 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.0 | 0.1 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.0 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.0 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 0.0 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.0 | 0.3 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.0 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.1 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.0 | GO:2000397 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.1 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.1 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.0 | 0.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.1 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.0 | 0.0 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.0 | 0.0 | GO:0003417 | growth plate cartilage development(GO:0003417) cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.0 | 0.1 | GO:0034398 | telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.0 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.0 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.0 | 0.1 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.1 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.1 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.1 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.0 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 0.0 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 0.7 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 0.1 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.0 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.1 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 0.1 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.0 | GO:0061582 | intestinal epithelial cell migration(GO:0061582) |
| 0.0 | 0.0 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.0 | 0.1 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.0 | 0.1 | GO:2000553 | positive regulation of T-helper 2 cell cytokine production(GO:2000553) |
| 0.0 | 0.1 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.7 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.1 | 0.4 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.4 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.2 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.2 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.2 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.0 | 0.1 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.0 | GO:0044753 | amphisome(GO:0044753) |
| 0.0 | 0.0 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.0 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.0 | 0.1 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.3 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.2 | GO:0035859 | Seh1-associated complex(GO:0035859) |
| 0.0 | 0.0 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.0 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.0 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.3 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.0 | 0.2 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.0 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.0 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.1 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.1 | 0.3 | GO:0032440 | 2-alkenal reductase [NAD(P)] activity(GO:0032440) |
| 0.1 | 0.9 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 0.3 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.1 | 0.3 | GO:0031071 | cysteine desulfurase activity(GO:0031071) |
| 0.1 | 0.3 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.1 | 0.3 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.1 | 0.3 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.3 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.1 | 0.3 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 0.7 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.3 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.1 | 0.2 | GO:0052830 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) |
| 0.0 | 0.1 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 0.2 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.4 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.2 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.0 | 0.3 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.6 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.2 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.2 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.2 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.0 | 0.4 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.1 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.0 | 0.2 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.4 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0004603 | phenylethanolamine N-methyltransferase activity(GO:0004603) |
| 0.0 | 0.8 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0033981 | D-dopachrome decarboxylase activity(GO:0033981) |
| 0.0 | 0.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.2 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.0 | 0.1 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.1 | GO:0050421 | cystathionine beta-synthase activity(GO:0004122) oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) nitrite reductase (NO-forming) activity(GO:0050421) carbon monoxide binding(GO:0070025) nitrite reductase activity(GO:0098809) |
| 0.0 | 0.1 | GO:0019107 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.0 | 0.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.3 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.0 | GO:0016434 | rRNA methyltransferase activity(GO:0008649) rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.1 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.0 | 0.1 | GO:0001512 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 0.0 | 0.1 | GO:0070260 | tyrosyl-RNA phosphodiesterase activity(GO:0036317) 5'-tyrosyl-DNA phosphodiesterase activity(GO:0070260) |
| 0.0 | 0.1 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.0 | 0.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.0 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0004532 | 3'-5'-exoribonuclease activity(GO:0000175) exoribonuclease activity(GO:0004532) exonuclease activity, active with either ribo- or deoxyribonucleic acids and producing 5'-phosphomonoesters(GO:0016796) exoribonuclease activity, producing 5'-phosphomonoesters(GO:0016896) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.0 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.1 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.0 | GO:0008478 | pyridoxal kinase activity(GO:0008478) lithium ion binding(GO:0031403) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.0 | 0.1 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.0 | 0.0 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.0 | GO:0031177 | acyl binding(GO:0000035) phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.0 | GO:0034038 | deoxyhypusine synthase activity(GO:0034038) |
| 0.0 | 0.3 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.0 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.2 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.0 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.3 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.4 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.0 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.0 | 0.0 | GO:0000384 | first spliceosomal transesterification activity(GO:0000384) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.0 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.1 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.0 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.1 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.0 | 0.0 | GO:0043734 | DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.0 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.1 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.2 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.9 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.7 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.9 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 0.3 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.2 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |