Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
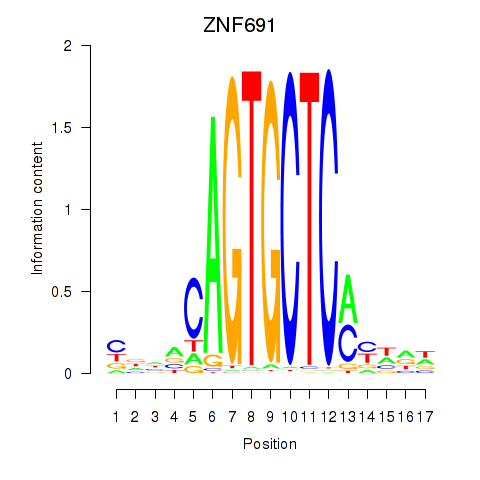
Results for ZNF691
Z-value: 0.70
Transcription factors associated with ZNF691
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF691
|
ENSG00000164011.13 | ZNF691 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF691 | hg19_v2_chr1_+_43312258_43312310 | -0.16 | 7.0e-01 | Click! |
Activity profile of ZNF691 motif
Sorted Z-values of ZNF691 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF691
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_147232669 | 1.69 |
ENST00000369237.1 |
GJA5 |
gap junction protein, alpha 5, 40kDa |
| chr16_+_57662419 | 0.83 |
ENST00000388812.4 ENST00000538815.1 ENST00000456916.1 ENST00000567154.1 ENST00000388813.5 ENST00000562558.1 ENST00000566271.2 |
GPR56 |
G protein-coupled receptor 56 |
| chr1_+_26856236 | 0.71 |
ENST00000374168.2 ENST00000374166.4 |
RPS6KA1 |
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chr16_+_57662138 | 0.69 |
ENST00000562414.1 ENST00000561969.1 ENST00000562631.1 ENST00000563445.1 ENST00000565338.1 ENST00000567702.1 |
GPR56 |
G protein-coupled receptor 56 |
| chr2_+_68961934 | 0.62 |
ENST00000409202.3 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr2_+_68961905 | 0.61 |
ENST00000295381.3 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr18_+_11751493 | 0.55 |
ENST00000269162.5 |
GNAL |
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr2_-_74669009 | 0.55 |
ENST00000272430.5 |
RTKN |
rhotekin |
| chr19_+_50084561 | 0.54 |
ENST00000246794.5 |
PRRG2 |
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr2_+_106468204 | 0.52 |
ENST00000425756.1 ENST00000393349.2 |
NCK2 |
NCK adaptor protein 2 |
| chr11_-_5323226 | 0.47 |
ENST00000380224.1 |
OR51B4 |
olfactory receptor, family 51, subfamily B, member 4 |
| chr19_-_51472222 | 0.43 |
ENST00000376851.3 |
KLK6 |
kallikrein-related peptidase 6 |
| chr16_+_53738053 | 0.42 |
ENST00000394647.3 |
FTO |
fat mass and obesity associated |
| chr19_-_51472031 | 0.40 |
ENST00000391808.1 |
KLK6 |
kallikrein-related peptidase 6 |
| chr8_+_32579341 | 0.39 |
ENST00000519240.1 ENST00000539990.1 |
NRG1 |
neuregulin 1 |
| chr14_-_92333873 | 0.39 |
ENST00000435962.2 |
TC2N |
tandem C2 domains, nuclear |
| chr2_+_68962014 | 0.35 |
ENST00000467265.1 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr11_+_121461097 | 0.35 |
ENST00000527934.1 |
SORL1 |
sortilin-related receptor, L(DLR class) A repeats containing |
| chr9_+_71944241 | 0.30 |
ENST00000257515.8 |
FAM189A2 |
family with sequence similarity 189, member A2 |
| chr5_-_35230434 | 0.27 |
ENST00000504500.1 |
PRLR |
prolactin receptor |
| chr11_-_36619771 | 0.26 |
ENST00000311485.3 ENST00000527033.1 ENST00000532616.1 |
RAG2 |
recombination activating gene 2 |
| chrX_+_102965835 | 0.24 |
ENST00000319560.6 |
TMEM31 |
transmembrane protein 31 |
| chrX_-_103268259 | 0.22 |
ENST00000217926.5 |
H2BFWT |
H2B histone family, member W, testis-specific |
| chrX_+_102469997 | 0.20 |
ENST00000372695.5 ENST00000372691.3 |
BEX4 |
brain expressed, X-linked 4 |
| chr15_-_83018198 | 0.20 |
ENST00000557886.1 |
RP13-996F3.4 |
golgin A6 family-like 19 |
| chr3_+_46412345 | 0.20 |
ENST00000292303.4 |
CCR5 |
chemokine (C-C motif) receptor 5 (gene/pseudogene) |
| chr16_+_84328429 | 0.19 |
ENST00000568638.1 |
WFDC1 |
WAP four-disulfide core domain 1 |
| chr17_+_30771279 | 0.19 |
ENST00000261712.3 ENST00000578213.1 ENST00000457654.2 ENST00000579451.1 |
PSMD11 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr1_-_203055129 | 0.19 |
ENST00000241651.4 |
MYOG |
myogenin (myogenic factor 4) |
| chr16_+_84328252 | 0.19 |
ENST00000219454.5 |
WFDC1 |
WAP four-disulfide core domain 1 |
| chr3_-_10452359 | 0.18 |
ENST00000452124.1 |
ATP2B2 |
ATPase, Ca++ transporting, plasma membrane 2 |
| chr9_+_139847347 | 0.17 |
ENST00000371632.3 |
LCN12 |
lipocalin 12 |
| chr17_-_41738931 | 0.16 |
ENST00000329168.3 ENST00000549132.1 |
MEOX1 |
mesenchyme homeobox 1 |
| chr4_-_83719983 | 0.16 |
ENST00000319540.4 |
SCD5 |
stearoyl-CoA desaturase 5 |
| chr8_-_145159083 | 0.15 |
ENST00000398712.2 |
SHARPIN |
SHANK-associated RH domain interactor |
| chr15_-_26108355 | 0.15 |
ENST00000356865.6 |
ATP10A |
ATPase, class V, type 10A |
| chr6_+_28493753 | 0.14 |
ENST00000469384.1 |
GPX5 |
glutathione peroxidase 5 (epididymal androgen-related protein) |
| chr15_+_84908573 | 0.14 |
ENST00000424966.1 ENST00000422563.2 |
GOLGA6L4 |
golgin A6 family-like 4 |
| chr17_-_73840415 | 0.14 |
ENST00000592386.1 ENST00000412096.2 ENST00000586147.1 |
UNC13D |
unc-13 homolog D (C. elegans) |
| chr4_-_120243545 | 0.13 |
ENST00000274024.3 |
FABP2 |
fatty acid binding protein 2, intestinal |
| chr20_-_1317555 | 0.13 |
ENST00000537552.1 |
AL136531.1 |
HCG2043693; Uncharacterized protein |
| chr9_+_84603687 | 0.13 |
ENST00000344803.2 |
SPATA31D1 |
SPATA31 subfamily D, member 1 |
| chr12_-_109221160 | 0.13 |
ENST00000326470.5 |
SSH1 |
slingshot protein phosphatase 1 |
| chr11_+_102188272 | 0.13 |
ENST00000532808.1 |
BIRC3 |
baculoviral IAP repeat containing 3 |
| chr9_+_139553306 | 0.13 |
ENST00000371699.1 |
EGFL7 |
EGF-like-domain, multiple 7 |
| chr12_-_51717875 | 0.13 |
ENST00000604560.1 |
BIN2 |
bridging integrator 2 |
| chr12_-_51717948 | 0.12 |
ENST00000267012.4 |
BIN2 |
bridging integrator 2 |
| chr8_+_145294015 | 0.12 |
ENST00000544576.1 |
MROH1 |
maestro heat-like repeat family member 1 |
| chr6_+_151186554 | 0.12 |
ENST00000367321.3 ENST00000367307.4 |
MTHFD1L |
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr11_-_57148619 | 0.12 |
ENST00000287143.2 |
PRG3 |
proteoglycan 3 |
| chr12_-_51718436 | 0.12 |
ENST00000544402.1 |
BIN2 |
bridging integrator 2 |
| chr11_+_33902189 | 0.12 |
ENST00000330381.2 |
AC132216.1 |
HCG1785179; PRO1787; Uncharacterized protein |
| chr3_-_46037299 | 0.11 |
ENST00000296137.2 |
FYCO1 |
FYVE and coiled-coil domain containing 1 |
| chr3_-_167452262 | 0.11 |
ENST00000487947.2 |
PDCD10 |
programmed cell death 10 |
| chr12_-_51717922 | 0.11 |
ENST00000452142.2 |
BIN2 |
bridging integrator 2 |
| chr5_-_39074479 | 0.10 |
ENST00000514735.1 ENST00000296782.5 ENST00000357387.3 |
RICTOR |
RPTOR independent companion of MTOR, complex 2 |
| chr1_+_248201474 | 0.10 |
ENST00000366479.2 |
OR2L2 |
olfactory receptor, family 2, subfamily L, member 2 |
| chr15_-_58571445 | 0.10 |
ENST00000558231.1 |
ALDH1A2 |
aldehyde dehydrogenase 1 family, member A2 |
| chr9_+_37485932 | 0.09 |
ENST00000377798.4 ENST00000442009.2 |
POLR1E |
polymerase (RNA) I polypeptide E, 53kDa |
| chr1_-_935361 | 0.09 |
ENST00000484667.2 |
HES4 |
hes family bHLH transcription factor 4 |
| chr17_-_38821373 | 0.09 |
ENST00000394052.3 |
KRT222 |
keratin 222 |
| chr11_+_5410607 | 0.09 |
ENST00000328611.3 |
OR51M1 |
olfactory receptor, family 51, subfamily M, member 1 |
| chr9_+_138453595 | 0.09 |
ENST00000479141.1 ENST00000371766.2 ENST00000277508.5 ENST00000433563.1 |
PAEP |
progestagen-associated endometrial protein |
| chr9_+_37486005 | 0.09 |
ENST00000377792.3 |
POLR1E |
polymerase (RNA) I polypeptide E, 53kDa |
| chr1_+_178482262 | 0.08 |
ENST00000367641.3 ENST00000367639.1 |
TEX35 |
testis expressed 35 |
| chr3_-_167452298 | 0.08 |
ENST00000475915.2 ENST00000462725.2 ENST00000461494.1 |
PDCD10 |
programmed cell death 10 |
| chr17_-_3417062 | 0.08 |
ENST00000570318.1 ENST00000541913.1 |
SPATA22 |
spermatogenesis associated 22 |
| chr22_-_43539346 | 0.08 |
ENST00000327555.5 ENST00000290429.6 |
MCAT |
malonyl CoA:ACP acyltransferase (mitochondrial) |
| chr9_+_135854091 | 0.08 |
ENST00000450530.1 ENST00000534944.1 |
GFI1B |
growth factor independent 1B transcription repressor |
| chr15_-_86338100 | 0.07 |
ENST00000536947.1 |
KLHL25 |
kelch-like family member 25 |
| chr10_+_104678102 | 0.07 |
ENST00000433628.2 |
CNNM2 |
cyclin M2 |
| chr12_-_11062161 | 0.07 |
ENST00000390677.2 |
TAS2R13 |
taste receptor, type 2, member 13 |
| chr12_-_23737534 | 0.07 |
ENST00000396007.2 |
SOX5 |
SRY (sex determining region Y)-box 5 |
| chr10_+_104678032 | 0.07 |
ENST00000369878.4 ENST00000369875.3 |
CNNM2 |
cyclin M2 |
| chr13_-_52027134 | 0.07 |
ENST00000311234.4 ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6 |
integrator complex subunit 6 |
| chr11_-_133826852 | 0.07 |
ENST00000533871.2 ENST00000321016.8 |
IGSF9B |
immunoglobulin superfamily, member 9B |
| chr3_-_167452614 | 0.06 |
ENST00000392750.2 ENST00000464360.1 ENST00000492139.1 ENST00000471885.1 ENST00000470131.1 |
PDCD10 |
programmed cell death 10 |
| chr2_+_202098203 | 0.06 |
ENST00000450491.1 ENST00000440732.1 ENST00000392258.3 |
CASP8 |
caspase 8, apoptosis-related cysteine peptidase |
| chr4_+_48492269 | 0.06 |
ENST00000327939.4 |
ZAR1 |
zygote arrest 1 |
| chr6_-_90529418 | 0.06 |
ENST00000439638.1 ENST00000369393.3 ENST00000428876.1 |
MDN1 |
MDN1, midasin homolog (yeast) |
| chr6_-_27279949 | 0.06 |
ENST00000444565.1 ENST00000377451.2 |
POM121L2 |
POM121 transmembrane nucleoporin-like 2 |
| chr5_+_7396141 | 0.06 |
ENST00000338316.4 |
ADCY2 |
adenylate cyclase 2 (brain) |
| chr1_+_33938236 | 0.05 |
ENST00000361328.3 ENST00000373413.2 |
ZSCAN20 |
zinc finger and SCAN domain containing 20 |
| chr19_+_40877583 | 0.05 |
ENST00000596470.1 |
PLD3 |
phospholipase D family, member 3 |
| chr1_+_178482212 | 0.05 |
ENST00000319416.2 ENST00000258298.2 ENST00000367643.3 ENST00000367642.3 |
TEX35 |
testis expressed 35 |
| chr17_+_80416050 | 0.05 |
ENST00000579198.1 ENST00000390006.4 ENST00000580296.1 |
NARF |
nuclear prelamin A recognition factor |
| chr6_+_31553978 | 0.05 |
ENST00000376096.1 ENST00000376099.1 ENST00000376110.3 |
LST1 |
leukocyte specific transcript 1 |
| chr10_-_61122220 | 0.05 |
ENST00000422313.2 ENST00000435852.2 ENST00000442566.3 ENST00000373868.2 ENST00000277705.6 ENST00000373867.3 ENST00000419214.2 |
FAM13C |
family with sequence similarity 13, member C |
| chr8_+_20831460 | 0.04 |
ENST00000522604.1 |
RP11-421P23.1 |
RP11-421P23.1 |
| chr5_-_131892501 | 0.04 |
ENST00000450655.1 |
IL5 |
interleukin 5 (colony-stimulating factor, eosinophil) |
| chr6_+_31553901 | 0.04 |
ENST00000418507.2 ENST00000438075.2 ENST00000376100.3 ENST00000376111.4 |
LST1 |
leukocyte specific transcript 1 |
| chr9_-_135754164 | 0.04 |
ENST00000298545.3 |
AK8 |
adenylate kinase 8 |
| chr15_+_84904525 | 0.04 |
ENST00000510439.2 |
GOLGA6L4 |
golgin A6 family-like 4 |
| chr1_+_15986364 | 0.04 |
ENST00000345034.1 |
RSC1A1 |
regulatory solute carrier protein, family 1, member 1 |
| chr10_+_122357721 | 0.04 |
ENST00000369071.2 |
C10orf85 |
chromosome 10 open reading frame 85 |
| chr7_-_143059780 | 0.03 |
ENST00000409578.1 ENST00000409346.1 |
FAM131B |
family with sequence similarity 131, member B |
| chr6_+_28493666 | 0.03 |
ENST00000412168.2 |
GPX5 |
glutathione peroxidase 5 (epididymal androgen-related protein) |
| chr6_+_31021973 | 0.03 |
ENST00000570223.1 ENST00000566475.1 ENST00000426185.1 |
HCG22 |
HLA complex group 22 |
| chr1_-_148202536 | 0.03 |
ENST00000544708.1 |
PPIAL4D |
peptidylprolyl isomerase A (cyclophilin A)-like 4D |
| chr8_+_140943416 | 0.03 |
ENST00000507535.3 |
C8orf17 |
chromosome 8 open reading frame 17 |
| chr5_-_175395283 | 0.03 |
ENST00000513482.1 ENST00000265097.4 |
THOC3 |
THO complex 3 |
| chrX_+_107069063 | 0.03 |
ENST00000262843.6 |
MID2 |
midline 2 |
| chr11_+_44748361 | 0.03 |
ENST00000533202.1 ENST00000533080.1 ENST00000520358.2 ENST00000520999.2 |
TSPAN18 |
tetraspanin 18 |
| chr17_-_61777459 | 0.03 |
ENST00000578993.1 ENST00000583211.1 ENST00000259006.3 |
LIMD2 |
LIM domain containing 2 |
| chr10_-_99258135 | 0.03 |
ENST00000327238.10 ENST00000327277.7 ENST00000355839.6 ENST00000437002.1 ENST00000422685.1 |
MMS19 |
MMS19 nucleotide excision repair homolog (S. cerevisiae) |
| chr17_+_37824700 | 0.03 |
ENST00000581428.1 |
PNMT |
phenylethanolamine N-methyltransferase |
| chr17_+_43922241 | 0.03 |
ENST00000329196.5 |
SPPL2C |
signal peptide peptidase like 2C |
| chr8_-_28347737 | 0.02 |
ENST00000517673.1 ENST00000518734.1 ENST00000346498.2 ENST00000380254.2 |
FBXO16 |
F-box protein 16 |
| chr1_+_26644441 | 0.02 |
ENST00000374213.2 |
CD52 |
CD52 molecule |
| chr9_+_116267536 | 0.02 |
ENST00000374136.1 |
RGS3 |
regulator of G-protein signaling 3 |
| chr17_+_20978854 | 0.02 |
ENST00000456235.1 |
AC087393.1 |
AC087393.1 |
| chr1_-_16763911 | 0.02 |
ENST00000375577.1 ENST00000335496.1 |
SPATA21 |
spermatogenesis associated 21 |
| chr4_-_7105056 | 0.02 |
ENST00000504402.1 ENST00000499242.2 ENST00000501888.2 |
RP11-367J11.3 |
RP11-367J11.3 |
| chr1_-_155232221 | 0.01 |
ENST00000355379.3 |
SCAMP3 |
secretory carrier membrane protein 3 |
| chr7_-_84569561 | 0.01 |
ENST00000439105.1 |
AC074183.4 |
AC074183.4 |
| chr6_-_144329531 | 0.01 |
ENST00000429150.1 ENST00000392309.1 ENST00000416623.1 ENST00000392307.1 |
PLAGL1 |
pleiomorphic adenoma gene-like 1 |
| chr1_-_111174054 | 0.01 |
ENST00000369770.3 |
KCNA2 |
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr2_-_203736452 | 0.01 |
ENST00000419460.1 |
ICA1L |
islet cell autoantigen 1,69kDa-like |
| chr15_+_89402148 | 0.01 |
ENST00000560601.1 |
ACAN |
aggrecan |
| chr19_-_15560730 | 0.00 |
ENST00000389282.4 ENST00000263381.7 |
WIZ |
widely interspaced zinc finger motifs |
| chr16_+_78133293 | 0.00 |
ENST00000566780.1 |
WWOX |
WW domain containing oxidoreductase |
| chr8_-_102181718 | 0.00 |
ENST00000565617.1 |
KB-1460A1.5 |
KB-1460A1.5 |
| chr8_-_121457332 | 0.00 |
ENST00000518918.1 |
MRPL13 |
mitochondrial ribosomal protein L13 |
| chr6_+_31021225 | 0.00 |
ENST00000565192.1 ENST00000562344.1 |
HCG22 |
HLA complex group 22 |
| chr22_+_23264766 | 0.00 |
ENST00000390331.2 |
IGLC7 |
immunoglobulin lambda constant 7 |
| chr14_-_74296806 | 0.00 |
ENST00000555539.1 |
RP5-1021I20.2 |
RP5-1021I20.2 |
| chr17_+_80416482 | 0.00 |
ENST00000309794.11 ENST00000345415.7 ENST00000457415.3 ENST00000584411.1 ENST00000412079.2 ENST00000577432.1 |
NARF |
nuclear prelamin A recognition factor |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0010652 | pulmonary valve formation(GO:0003193) regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) foramen ovale closure(GO:0035922) regulation of bundle of His cell action potential(GO:0098905) |
| 0.1 | 0.4 | GO:0035552 | oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.1 | 0.3 | GO:1902948 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.1 | 0.7 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 | 0.5 | GO:0032185 | septin cytoskeleton organization(GO:0032185) |
| 0.1 | 0.5 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.1 | 0.3 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.1 | 0.4 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.1 | 0.2 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.3 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.2 | GO:0001189 | RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.0 | 0.3 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 | 0.1 | GO:1904719 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.1 | GO:0001694 | histamine biosynthetic process(GO:0001694) |
| 0.0 | 0.8 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.2 | GO:1903862 | regulation of muscle atrophy(GO:0014735) response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.1 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.0 | 0.1 | GO:1902490 | regulation of sperm capacitation(GO:1902490) |
| 0.0 | 0.2 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.2 | GO:0001757 | somite specification(GO:0001757) sclerotome development(GO:0061056) |
| 0.0 | 0.2 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.1 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.0 | 0.5 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.0 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.0 | 0.1 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.3 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.1 | GO:0070424 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0035799 | ureter maturation(GO:0035799) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.1 | 0.4 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.1 | 0.3 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.4 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.2 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.0 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.0 | 0.3 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.8 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.5 | PID REELIN PATHWAY | Reelin signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.2 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.4 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.7 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |