Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
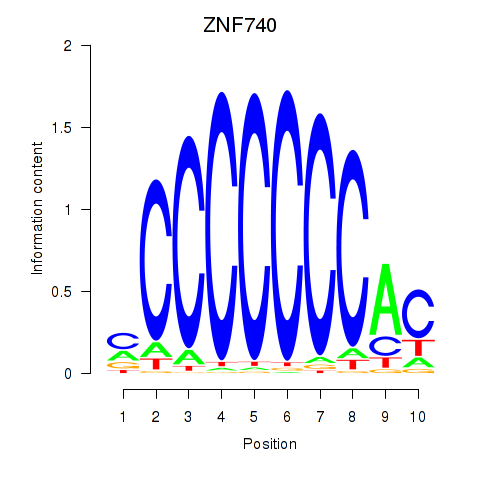
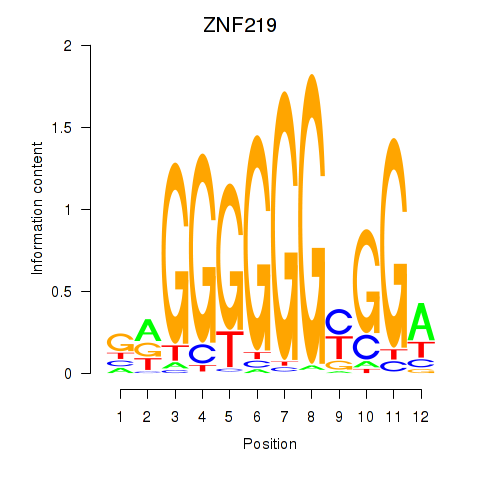
Results for ZNF740_ZNF219
Z-value: 0.85
Transcription factors associated with ZNF740_ZNF219
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF740
|
ENSG00000139651.9 | ZNF740 |
|
ZNF219
|
ENSG00000165804.11 | ZNF219 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF219 | hg19_v2_chr14_-_21562648_21562659 | 0.98 | 3.4e-05 | Click! |
| ZNF740 | hg19_v2_chr12_+_53574464_53574539 | 0.92 | 1.3e-03 | Click! |
Activity profile of ZNF740_ZNF219 motif
Sorted Z-values of ZNF740_ZNF219 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF740_ZNF219
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_95653302 | 2.69 |
ENST00000423620.2 ENST00000433389.2 |
ESRP1 |
epithelial splicing regulatory protein 1 |
| chr8_+_95653427 | 2.68 |
ENST00000454170.2 |
ESRP1 |
epithelial splicing regulatory protein 1 |
| chr8_+_95653373 | 2.66 |
ENST00000358397.5 |
ESRP1 |
epithelial splicing regulatory protein 1 |
| chr20_+_58179582 | 1.47 |
ENST00000371015.1 ENST00000395639.4 |
PHACTR3 |
phosphatase and actin regulator 3 |
| chr6_-_32157947 | 1.20 |
ENST00000375050.4 |
PBX2 |
pre-B-cell leukemia homeobox 2 |
| chr12_-_54785054 | 0.83 |
ENST00000352268.6 ENST00000549962.1 |
ZNF385A |
zinc finger protein 385A |
| chr20_-_22565101 | 0.82 |
ENST00000419308.2 |
FOXA2 |
forkhead box A2 |
| chr3_+_189507432 | 0.82 |
ENST00000354600.5 |
TP63 |
tumor protein p63 |
| chr1_-_242687989 | 0.75 |
ENST00000442594.2 |
PLD5 |
phospholipase D family, member 5 |
| chr6_-_33285505 | 0.68 |
ENST00000431845.2 |
ZBTB22 |
zinc finger and BTB domain containing 22 |
| chr11_-_63933504 | 0.64 |
ENST00000255681.6 |
MACROD1 |
MACRO domain containing 1 |
| chr16_+_81478775 | 0.60 |
ENST00000537098.3 |
CMIP |
c-Maf inducing protein |
| chr14_+_93897272 | 0.60 |
ENST00000393151.2 |
UNC79 |
unc-79 homolog (C. elegans) |
| chr17_+_7788104 | 0.53 |
ENST00000380358.4 |
CHD3 |
chromodomain helicase DNA binding protein 3 |
| chr1_+_156030937 | 0.53 |
ENST00000361084.5 |
RAB25 |
RAB25, member RAS oncogene family |
| chr3_-_171178157 | 0.52 |
ENST00000465393.1 ENST00000436636.2 ENST00000369326.5 ENST00000538048.1 ENST00000341852.6 |
TNIK |
TRAF2 and NCK interacting kinase |
| chr12_-_53614155 | 0.48 |
ENST00000543726.1 |
RARG |
retinoic acid receptor, gamma |
| chr1_-_19283163 | 0.45 |
ENST00000455833.2 |
IFFO2 |
intermediate filament family orphan 2 |
| chr20_-_50808236 | 0.45 |
ENST00000361387.2 |
ZFP64 |
ZFP64 zinc finger protein |
| chr1_+_26856236 | 0.44 |
ENST00000374168.2 ENST00000374166.4 |
RPS6KA1 |
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chr14_-_105635090 | 0.44 |
ENST00000331782.3 ENST00000347004.2 |
JAG2 |
jagged 2 |
| chr3_-_171177852 | 0.40 |
ENST00000284483.8 ENST00000475336.1 ENST00000357327.5 ENST00000460047.1 ENST00000488470.1 ENST00000470834.1 |
TNIK |
TRAF2 and NCK interacting kinase |
| chrX_-_128788914 | 0.39 |
ENST00000429967.1 ENST00000307484.6 |
APLN |
apelin |
| chr14_+_75745477 | 0.39 |
ENST00000303562.4 ENST00000554617.1 ENST00000554212.1 ENST00000535987.1 ENST00000555242.1 |
FOS |
FBJ murine osteosarcoma viral oncogene homolog |
| chr17_-_39674668 | 0.38 |
ENST00000393981.3 |
KRT15 |
keratin 15 |
| chr3_+_189507523 | 0.38 |
ENST00000437221.1 ENST00000392463.2 ENST00000392461.3 ENST00000449992.1 ENST00000456148.1 |
TP63 |
tumor protein p63 |
| chr9_-_124990680 | 0.38 |
ENST00000541397.2 ENST00000560485.1 |
LHX6 |
LIM homeobox 6 |
| chr9_-_124991124 | 0.38 |
ENST00000394319.4 ENST00000340587.3 |
LHX6 |
LIM homeobox 6 |
| chr11_-_46142505 | 0.37 |
ENST00000524497.1 ENST00000418153.2 |
PHF21A |
PHD finger protein 21A |
| chr6_-_34664612 | 0.36 |
ENST00000374023.3 ENST00000374026.3 |
C6orf106 |
chromosome 6 open reading frame 106 |
| chr14_-_21994525 | 0.36 |
ENST00000538754.1 |
SALL2 |
spalt-like transcription factor 2 |
| chr17_-_42277203 | 0.36 |
ENST00000587097.1 |
ATXN7L3 |
ataxin 7-like 3 |
| chr12_-_53614043 | 0.36 |
ENST00000338561.5 |
RARG |
retinoic acid receptor, gamma |
| chr1_-_33430286 | 0.36 |
ENST00000373456.7 ENST00000356990.5 ENST00000235150.4 |
RNF19B |
ring finger protein 19B |
| chr7_+_69064300 | 0.35 |
ENST00000342771.4 |
AUTS2 |
autism susceptibility candidate 2 |
| chr19_+_45754505 | 0.34 |
ENST00000262891.4 ENST00000300843.4 |
MARK4 |
MAP/microtubule affinity-regulating kinase 4 |
| chr6_+_17281573 | 0.33 |
ENST00000379052.5 |
RBM24 |
RNA binding motif protein 24 |
| chr19_+_34287174 | 0.33 |
ENST00000587559.1 ENST00000588637.1 |
KCTD15 |
potassium channel tetramerization domain containing 15 |
| chr15_-_88799384 | 0.33 |
ENST00000540489.2 ENST00000557856.1 ENST00000558676.1 |
NTRK3 |
neurotrophic tyrosine kinase, receptor, type 3 |
| chr20_-_31071239 | 0.32 |
ENST00000359676.5 |
C20orf112 |
chromosome 20 open reading frame 112 |
| chr10_+_111967345 | 0.31 |
ENST00000332674.5 ENST00000453116.1 |
MXI1 |
MAX interactor 1, dimerization protein |
| chr11_-_46142948 | 0.31 |
ENST00000257821.4 |
PHF21A |
PHD finger protein 21A |
| chr1_-_6321035 | 0.31 |
ENST00000377893.2 |
GPR153 |
G protein-coupled receptor 153 |
| chr1_-_41131326 | 0.30 |
ENST00000372684.3 |
RIMS3 |
regulating synaptic membrane exocytosis 3 |
| chr18_+_3449821 | 0.30 |
ENST00000407501.2 ENST00000405385.3 ENST00000546979.1 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr12_-_26278030 | 0.29 |
ENST00000242728.4 |
BHLHE41 |
basic helix-loop-helix family, member e41 |
| chr6_-_143266297 | 0.29 |
ENST00000367603.2 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
| chr7_+_21467642 | 0.29 |
ENST00000222584.3 ENST00000432066.2 |
SP4 |
Sp4 transcription factor |
| chr2_-_64371546 | 0.29 |
ENST00000358912.4 |
PELI1 |
pellino E3 ubiquitin protein ligase 1 |
| chr12_-_57522813 | 0.29 |
ENST00000556155.1 |
STAT6 |
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr17_+_55333876 | 0.28 |
ENST00000284073.2 |
MSI2 |
musashi RNA-binding protein 2 |
| chr9_-_23821273 | 0.28 |
ENST00000380110.4 |
ELAVL2 |
ELAV like neuron-specific RNA binding protein 2 |
| chr17_+_54671047 | 0.28 |
ENST00000332822.4 |
NOG |
noggin |
| chr18_+_56530136 | 0.28 |
ENST00000591083.1 |
ZNF532 |
zinc finger protein 532 |
| chr18_+_33877654 | 0.27 |
ENST00000257209.4 ENST00000445677.1 ENST00000590592.1 ENST00000359247.4 |
FHOD3 |
formin homology 2 domain containing 3 |
| chr12_-_54779511 | 0.26 |
ENST00000551109.1 ENST00000546970.1 |
ZNF385A |
zinc finger protein 385A |
| chr17_+_43239191 | 0.26 |
ENST00000589230.1 |
HEXIM2 |
hexamethylene bis-acetamide inducible 2 |
| chr7_-_105332084 | 0.26 |
ENST00000472195.1 |
ATXN7L1 |
ataxin 7-like 1 |
| chr7_+_153749732 | 0.26 |
ENST00000377770.3 |
DPP6 |
dipeptidyl-peptidase 6 |
| chr15_-_43212996 | 0.26 |
ENST00000567840.1 |
TTBK2 |
tau tubulin kinase 2 |
| chr8_-_494824 | 0.25 |
ENST00000427263.2 ENST00000324079.6 |
TDRP |
testis development related protein |
| chr19_-_38746979 | 0.25 |
ENST00000591291.1 |
PPP1R14A |
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr11_+_134201911 | 0.25 |
ENST00000389881.3 |
GLB1L2 |
galactosidase, beta 1-like 2 |
| chr1_-_242687676 | 0.25 |
ENST00000536534.2 |
PLD5 |
phospholipase D family, member 5 |
| chr15_-_43212836 | 0.24 |
ENST00000566931.1 ENST00000564431.1 ENST00000567274.1 |
TTBK2 |
tau tubulin kinase 2 |
| chr2_-_46769694 | 0.24 |
ENST00000522587.1 |
ATP6V1E2 |
ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E2 |
| chr14_-_99737565 | 0.24 |
ENST00000357195.3 |
BCL11B |
B-cell CLL/lymphoma 11B (zinc finger protein) |
| chr1_+_43148059 | 0.24 |
ENST00000321358.7 ENST00000332220.6 |
YBX1 |
Y box binding protein 1 |
| chr17_-_74533963 | 0.24 |
ENST00000293230.5 |
CYGB |
cytoglobin |
| chr17_-_6459802 | 0.24 |
ENST00000262483.8 |
PITPNM3 |
PITPNM family member 3 |
| chr17_-_27278445 | 0.23 |
ENST00000268756.3 ENST00000584685.1 |
PHF12 |
PHD finger protein 12 |
| chr11_-_46142615 | 0.23 |
ENST00000529734.1 ENST00000323180.6 |
PHF21A |
PHD finger protein 21A |
| chr19_+_11457232 | 0.23 |
ENST00000587531.1 |
CCDC159 |
coiled-coil domain containing 159 |
| chr1_+_13910757 | 0.22 |
ENST00000376061.4 ENST00000513143.1 |
PDPN |
podoplanin |
| chr12_-_54785074 | 0.22 |
ENST00000338010.5 ENST00000550774.1 |
ZNF385A |
zinc finger protein 385A |
| chr6_+_18155560 | 0.22 |
ENST00000546309.2 ENST00000388870.2 ENST00000397244.1 |
KDM1B |
lysine (K)-specific demethylase 1B |
| chr1_-_38273840 | 0.22 |
ENST00000373044.2 |
YRDC |
yrdC N(6)-threonylcarbamoyltransferase domain containing |
| chr14_-_21566731 | 0.22 |
ENST00000360947.3 |
ZNF219 |
zinc finger protein 219 |
| chr1_+_212458834 | 0.22 |
ENST00000261461.2 |
PPP2R5A |
protein phosphatase 2, regulatory subunit B', alpha |
| chr2_-_148779106 | 0.21 |
ENST00000416719.1 ENST00000264169.2 |
ORC4 |
origin recognition complex, subunit 4 |
| chr16_+_30935418 | 0.21 |
ENST00000338343.4 |
FBXL19 |
F-box and leucine-rich repeat protein 19 |
| chr22_-_39640756 | 0.21 |
ENST00000331163.6 |
PDGFB |
platelet-derived growth factor beta polypeptide |
| chrX_+_70316005 | 0.21 |
ENST00000374259.3 |
FOXO4 |
forkhead box O4 |
| chrX_+_152907913 | 0.21 |
ENST00000370167.4 |
DUSP9 |
dual specificity phosphatase 9 |
| chr22_+_46546494 | 0.21 |
ENST00000396000.2 ENST00000262735.5 ENST00000420804.1 |
PPARA |
peroxisome proliferator-activated receptor alpha |
| chr6_+_18155632 | 0.21 |
ENST00000297792.5 |
KDM1B |
lysine (K)-specific demethylase 1B |
| chr7_-_151217001 | 0.21 |
ENST00000262187.5 |
RHEB |
Ras homolog enriched in brain |
| chr13_-_41240717 | 0.20 |
ENST00000379561.5 |
FOXO1 |
forkhead box O1 |
| chr17_+_56160768 | 0.20 |
ENST00000579991.2 |
DYNLL2 |
dynein, light chain, LC8-type 2 |
| chr20_-_31071309 | 0.20 |
ENST00000326071.4 |
C20orf112 |
chromosome 20 open reading frame 112 |
| chr19_+_36208877 | 0.20 |
ENST00000420124.1 ENST00000222270.7 ENST00000341701.1 |
KMT2B |
Histone-lysine N-methyltransferase 2B |
| chr19_-_41222775 | 0.20 |
ENST00000324464.3 ENST00000450541.1 ENST00000594720.1 |
ADCK4 |
aarF domain containing kinase 4 |
| chr20_-_18038521 | 0.20 |
ENST00000278780.6 |
OVOL2 |
ovo-like zinc finger 2 |
| chr17_+_36861735 | 0.20 |
ENST00000378137.5 ENST00000325718.7 |
MLLT6 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6 |
| chr19_+_34287751 | 0.20 |
ENST00000590771.1 ENST00000589786.1 ENST00000284006.6 ENST00000588881.1 |
KCTD15 |
potassium channel tetramerization domain containing 15 |
| chr17_+_1646130 | 0.20 |
ENST00000453066.1 ENST00000324015.3 ENST00000450523.2 ENST00000453723.1 ENST00000382061.4 |
SERPINF2 |
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2 |
| chr2_-_208030647 | 0.19 |
ENST00000309446.6 |
KLF7 |
Kruppel-like factor 7 (ubiquitous) |
| chr17_+_43238438 | 0.19 |
ENST00000593138.1 ENST00000586681.1 |
HEXIM2 |
hexamethylene bis-acetamide inducible 2 |
| chr12_+_57482877 | 0.19 |
ENST00000342556.6 ENST00000357680.4 |
NAB2 |
NGFI-A binding protein 2 (EGR1 binding protein 2) |
| chr19_+_35759824 | 0.19 |
ENST00000343550.5 |
USF2 |
upstream transcription factor 2, c-fos interacting |
| chr12_+_58148842 | 0.19 |
ENST00000266643.5 |
MARCH9 |
membrane-associated ring finger (C3HC4) 9 |
| chr1_-_156051789 | 0.19 |
ENST00000532414.2 |
MEX3A |
mex-3 RNA binding family member A |
| chr20_+_35201857 | 0.19 |
ENST00000373874.2 |
TGIF2 |
TGFB-induced factor homeobox 2 |
| chr10_+_120967072 | 0.19 |
ENST00000392870.2 |
GRK5 |
G protein-coupled receptor kinase 5 |
| chr17_+_57297807 | 0.18 |
ENST00000284116.4 ENST00000581140.1 ENST00000581276.1 |
GDPD1 |
glycerophosphodiester phosphodiesterase domain containing 1 |
| chr4_+_146403912 | 0.18 |
ENST00000507367.1 ENST00000394092.2 ENST00000515385.1 |
SMAD1 |
SMAD family member 1 |
| chr16_+_23847267 | 0.18 |
ENST00000321728.7 |
PRKCB |
protein kinase C, beta |
| chr10_-_13390021 | 0.18 |
ENST00000537130.1 |
SEPHS1 |
selenophosphate synthetase 1 |
| chr1_-_244013384 | 0.18 |
ENST00000366539.1 |
AKT3 |
v-akt murine thymoma viral oncogene homolog 3 |
| chr1_+_6845578 | 0.18 |
ENST00000467404.2 ENST00000439411.2 |
CAMTA1 |
calmodulin binding transcription activator 1 |
| chr19_+_47105309 | 0.18 |
ENST00000599839.1 ENST00000596362.1 |
CALM3 |
calmodulin 3 (phosphorylase kinase, delta) |
| chr14_-_99737822 | 0.18 |
ENST00000345514.2 ENST00000443726.2 |
BCL11B |
B-cell CLL/lymphoma 11B (zinc finger protein) |
| chr18_+_21594585 | 0.18 |
ENST00000317571.3 |
TTC39C |
tetratricopeptide repeat domain 39C |
| chr20_+_35201993 | 0.18 |
ENST00000373872.4 |
TGIF2 |
TGFB-induced factor homeobox 2 |
| chr6_+_43739697 | 0.17 |
ENST00000230480.6 |
VEGFA |
vascular endothelial growth factor A |
| chr6_-_42419649 | 0.17 |
ENST00000372922.4 ENST00000541110.1 ENST00000372917.4 |
TRERF1 |
transcriptional regulating factor 1 |
| chr10_+_99332529 | 0.17 |
ENST00000455090.1 |
ANKRD2 |
ankyrin repeat domain 2 (stretch responsive muscle) |
| chr17_-_7154984 | 0.17 |
ENST00000574322.1 |
CTDNEP1 |
CTD nuclear envelope phosphatase 1 |
| chr3_-_129612394 | 0.17 |
ENST00000505616.1 ENST00000426664.2 |
TMCC1 |
transmembrane and coiled-coil domain family 1 |
| chr4_-_99850243 | 0.17 |
ENST00000280892.6 ENST00000511644.1 ENST00000504432.1 ENST00000505992.1 |
EIF4E |
eukaryotic translation initiation factor 4E |
| chr1_-_204121102 | 0.17 |
ENST00000367202.4 |
ETNK2 |
ethanolamine kinase 2 |
| chr19_-_46476791 | 0.17 |
ENST00000263257.5 |
NOVA2 |
neuro-oncological ventral antigen 2 |
| chr14_-_21493649 | 0.16 |
ENST00000553442.1 ENST00000555869.1 ENST00000556457.1 ENST00000397844.2 ENST00000554415.1 |
NDRG2 |
NDRG family member 2 |
| chr1_-_156470515 | 0.16 |
ENST00000340875.5 ENST00000368240.2 ENST00000353795.3 |
MEF2D |
myocyte enhancer factor 2D |
| chr1_+_211432700 | 0.16 |
ENST00000452621.2 |
RCOR3 |
REST corepressor 3 |
| chr8_+_123793633 | 0.16 |
ENST00000314393.4 |
ZHX2 |
zinc fingers and homeoboxes 2 |
| chr18_-_53255766 | 0.16 |
ENST00000566286.1 ENST00000564999.1 ENST00000566279.1 ENST00000354452.3 ENST00000356073.4 |
TCF4 |
transcription factor 4 |
| chr2_+_220325441 | 0.16 |
ENST00000396688.1 |
SPEG |
SPEG complex locus |
| chr2_-_64881018 | 0.16 |
ENST00000313349.3 |
SERTAD2 |
SERTA domain containing 2 |
| chr3_+_52017454 | 0.16 |
ENST00000476854.1 ENST00000476351.1 ENST00000494103.1 ENST00000404366.2 ENST00000469863.1 |
ACY1 |
aminoacylase 1 |
| chr1_+_13910194 | 0.16 |
ENST00000376057.4 ENST00000510906.1 |
PDPN |
podoplanin |
| chr14_-_81687575 | 0.16 |
ENST00000434192.2 |
GTF2A1 |
general transcription factor IIA, 1, 19/37kDa |
| chr2_-_217236750 | 0.16 |
ENST00000273067.4 |
MARCH4 |
membrane-associated ring finger (C3HC4) 4, E3 ubiquitin protein ligase |
| chr11_-_5531215 | 0.16 |
ENST00000311659.4 |
UBQLN3 |
ubiquilin 3 |
| chr1_-_204121298 | 0.16 |
ENST00000367199.2 |
ETNK2 |
ethanolamine kinase 2 |
| chr10_-_86001210 | 0.16 |
ENST00000372105.3 |
LRIT1 |
leucine-rich repeat, immunoglobulin-like and transmembrane domains 1 |
| chr4_+_146402925 | 0.16 |
ENST00000302085.4 |
SMAD1 |
SMAD family member 1 |
| chr19_-_49015050 | 0.16 |
ENST00000600059.1 |
LMTK3 |
lemur tyrosine kinase 3 |
| chr15_-_68497657 | 0.16 |
ENST00000448060.2 ENST00000467889.1 |
CALML4 |
calmodulin-like 4 |
| chr16_+_11762270 | 0.16 |
ENST00000329565.5 |
SNN |
stannin |
| chr17_+_34136459 | 0.16 |
ENST00000588240.1 ENST00000590273.1 ENST00000588441.1 ENST00000587272.1 ENST00000592237.1 ENST00000311979.3 |
TAF15 |
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa |
| chr12_+_52445191 | 0.15 |
ENST00000243050.1 ENST00000394825.1 ENST00000550763.1 ENST00000394824.2 ENST00000548232.1 ENST00000562373.1 |
NR4A1 |
nuclear receptor subfamily 4, group A, member 1 |
| chr13_+_20532848 | 0.15 |
ENST00000382874.2 |
ZMYM2 |
zinc finger, MYM-type 2 |
| chr19_+_797392 | 0.15 |
ENST00000350092.4 ENST00000349038.4 ENST00000586481.1 ENST00000585535.1 |
PTBP1 |
polypyrimidine tract binding protein 1 |
| chr3_+_193853927 | 0.15 |
ENST00000232424.3 |
HES1 |
hes family bHLH transcription factor 1 |
| chr13_+_47127293 | 0.15 |
ENST00000311191.6 |
LRCH1 |
leucine-rich repeats and calponin homology (CH) domain containing 1 |
| chr17_-_7155274 | 0.15 |
ENST00000318988.6 ENST00000575783.1 ENST00000573600.1 |
CTDNEP1 |
CTD nuclear envelope phosphatase 1 |
| chr16_+_3074002 | 0.15 |
ENST00000326266.8 ENST00000574549.1 ENST00000575576.1 ENST00000253952.9 |
THOC6 |
THO complex 6 homolog (Drosophila) |
| chr1_-_204121013 | 0.15 |
ENST00000367201.3 |
ETNK2 |
ethanolamine kinase 2 |
| chr19_+_35759968 | 0.15 |
ENST00000222305.3 ENST00000595068.1 ENST00000379134.3 ENST00000594064.1 ENST00000598058.1 |
USF2 |
upstream transcription factor 2, c-fos interacting |
| chr14_-_21493884 | 0.15 |
ENST00000556974.1 ENST00000554419.1 ENST00000298687.5 ENST00000397858.1 ENST00000360463.3 ENST00000350792.3 ENST00000397847.2 |
NDRG2 |
NDRG family member 2 |
| chr3_-_133614297 | 0.15 |
ENST00000486858.1 ENST00000477759.1 |
RAB6B |
RAB6B, member RAS oncogene family |
| chr12_+_53773944 | 0.15 |
ENST00000551969.1 ENST00000327443.4 |
SP1 |
Sp1 transcription factor |
| chr8_+_104513086 | 0.15 |
ENST00000406091.3 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr19_-_18653781 | 0.15 |
ENST00000596558.2 ENST00000453489.2 |
FKBP8 |
FK506 binding protein 8, 38kDa |
| chr4_+_119810134 | 0.15 |
ENST00000434046.2 |
SYNPO2 |
synaptopodin 2 |
| chr6_+_37787704 | 0.14 |
ENST00000474522.1 |
ZFAND3 |
zinc finger, AN1-type domain 3 |
| chr14_+_65171099 | 0.14 |
ENST00000247226.7 |
PLEKHG3 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr16_+_57662138 | 0.14 |
ENST00000562414.1 ENST00000561969.1 ENST00000562631.1 ENST00000563445.1 ENST00000565338.1 ENST00000567702.1 |
GPR56 |
G protein-coupled receptor 56 |
| chr12_+_122064673 | 0.14 |
ENST00000537188.1 |
ORAI1 |
ORAI calcium release-activated calcium modulator 1 |
| chr8_+_109455845 | 0.14 |
ENST00000220853.3 |
EMC2 |
ER membrane protein complex subunit 2 |
| chr17_-_46688334 | 0.14 |
ENST00000239165.7 |
HOXB7 |
homeobox B7 |
| chr17_-_42143963 | 0.14 |
ENST00000585388.1 ENST00000293406.3 |
LSM12 |
LSM12 homolog (S. cerevisiae) |
| chr17_-_42200996 | 0.14 |
ENST00000587135.1 ENST00000225983.6 ENST00000393622.2 ENST00000588703.1 |
HDAC5 |
histone deacetylase 5 |
| chr9_+_116638562 | 0.14 |
ENST00000374126.5 ENST00000288466.7 |
ZNF618 |
zinc finger protein 618 |
| chr11_-_130184470 | 0.14 |
ENST00000357899.4 ENST00000397753.1 |
ZBTB44 |
zinc finger and BTB domain containing 44 |
| chr1_+_24117627 | 0.14 |
ENST00000400061.1 |
LYPLA2 |
lysophospholipase II |
| chrX_-_80065146 | 0.14 |
ENST00000373275.4 |
BRWD3 |
bromodomain and WD repeat domain containing 3 |
| chr5_-_134369973 | 0.14 |
ENST00000265340.7 |
PITX1 |
paired-like homeodomain 1 |
| chr1_+_161129254 | 0.13 |
ENST00000368002.3 ENST00000289865.8 ENST00000479344.1 ENST00000368001.1 |
USP21 |
ubiquitin specific peptidase 21 |
| chr20_-_52210368 | 0.13 |
ENST00000371471.2 |
ZNF217 |
zinc finger protein 217 |
| chr17_-_74533734 | 0.13 |
ENST00000589342.1 |
CYGB |
cytoglobin |
| chr12_+_53774423 | 0.13 |
ENST00000426431.2 |
SP1 |
Sp1 transcription factor |
| chr2_+_70142189 | 0.13 |
ENST00000264444.2 |
MXD1 |
MAX dimerization protein 1 |
| chr17_-_72869140 | 0.13 |
ENST00000583917.1 ENST00000293195.5 ENST00000442102.2 |
FDXR |
ferredoxin reductase |
| chr17_-_42200958 | 0.13 |
ENST00000336057.5 |
HDAC5 |
histone deacetylase 5 |
| chr14_-_23451845 | 0.13 |
ENST00000262713.2 |
AJUBA |
ajuba LIM protein |
| chr7_+_20370746 | 0.13 |
ENST00000222573.4 |
ITGB8 |
integrin, beta 8 |
| chr10_-_133795416 | 0.13 |
ENST00000540159.1 ENST00000368636.4 |
BNIP3 |
BCL2/adenovirus E1B 19kDa interacting protein 3 |
| chr19_+_7660716 | 0.13 |
ENST00000160298.4 ENST00000446248.2 |
CAMSAP3 |
calmodulin regulated spectrin-associated protein family, member 3 |
| chr1_+_2985760 | 0.13 |
ENST00000378391.2 ENST00000514189.1 ENST00000270722.5 |
PRDM16 |
PR domain containing 16 |
| chr19_-_1568057 | 0.13 |
ENST00000402693.4 ENST00000388824.6 |
MEX3D |
mex-3 RNA binding family member D |
| chr7_+_106809406 | 0.13 |
ENST00000468410.1 ENST00000478930.1 ENST00000464009.1 ENST00000222574.4 |
HBP1 |
HMG-box transcription factor 1 |
| chr22_+_46449674 | 0.13 |
ENST00000381051.2 |
FLJ27365 |
hsa-mir-4763 |
| chr8_+_124428959 | 0.13 |
ENST00000287387.2 ENST00000523984.1 |
WDYHV1 |
WDYHV motif containing 1 |
| chr18_-_19748331 | 0.13 |
ENST00000584201.1 |
GATA6-AS1 |
GATA6 antisense RNA 1 (head to head) |
| chr18_-_19748379 | 0.13 |
ENST00000579431.1 |
GATA6-AS1 |
GATA6 antisense RNA 1 (head to head) |
| chr1_+_24117693 | 0.12 |
ENST00000374503.3 ENST00000374502.3 |
LYPLA2 |
lysophospholipase II |
| chr6_+_130339710 | 0.12 |
ENST00000526087.1 ENST00000533560.1 ENST00000361794.2 |
L3MBTL3 |
l(3)mbt-like 3 (Drosophila) |
| chr16_+_57662419 | 0.12 |
ENST00000388812.4 ENST00000538815.1 ENST00000456916.1 ENST00000567154.1 ENST00000388813.5 ENST00000562558.1 ENST00000566271.2 |
GPR56 |
G protein-coupled receptor 56 |
| chr20_-_31124186 | 0.12 |
ENST00000375678.3 |
C20orf112 |
chromosome 20 open reading frame 112 |
| chr3_-_113415441 | 0.12 |
ENST00000491165.1 ENST00000316407.4 |
KIAA2018 |
KIAA2018 |
| chr2_+_24272543 | 0.12 |
ENST00000380991.4 |
FKBP1B |
FK506 binding protein 1B, 12.6 kDa |
| chr6_+_30850697 | 0.12 |
ENST00000509639.1 ENST00000412274.2 ENST00000507901.1 ENST00000507046.1 ENST00000437124.2 ENST00000454612.2 ENST00000396342.2 |
DDR1 |
discoidin domain receptor tyrosine kinase 1 |
| chr14_+_105190514 | 0.12 |
ENST00000330877.2 |
ADSSL1 |
adenylosuccinate synthase like 1 |
| chr11_+_64073022 | 0.12 |
ENST00000406310.1 ENST00000000442.6 ENST00000539594.1 |
ESRRA |
estrogen-related receptor alpha |
| chr22_+_29168652 | 0.12 |
ENST00000249064.4 ENST00000444523.1 ENST00000448492.2 ENST00000421503.2 |
CCDC117 |
coiled-coil domain containing 117 |
| chr2_+_220306745 | 0.12 |
ENST00000431523.1 ENST00000396698.1 ENST00000396695.2 |
SPEG |
SPEG complex locus |
| chr16_+_67063262 | 0.12 |
ENST00000565389.1 |
CBFB |
core-binding factor, beta subunit |
| chr14_+_96968707 | 0.12 |
ENST00000216277.8 ENST00000557320.1 ENST00000557471.1 |
PAPOLA |
poly(A) polymerase alpha |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) |
| 0.3 | 0.8 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.3 | 1.1 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.2 | 0.8 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.1 | 0.4 | GO:0019056 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.1 | 0.4 | GO:0043396 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.1 | 0.4 | GO:1904328 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.1 | 0.4 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.1 | 1.0 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.1 | 0.3 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) |
| 0.1 | 0.6 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.4 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.1 | 0.3 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.1 | 0.3 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.1 | 0.4 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.1 | 0.2 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 0.5 | GO:0036166 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.1 | 0.2 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.1 | 0.4 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 | 0.4 | GO:2000489 | regulation of hepatic stellate cell activation(GO:2000489) |
| 0.1 | 0.1 | GO:0051944 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.1 | 0.2 | GO:0071469 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) cellular response to alkaline pH(GO:0071469) |
| 0.1 | 0.2 | GO:1903570 | coronary vein morphogenesis(GO:0003169) regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.1 | 0.7 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 | 0.3 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.2 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.1 | 0.2 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.1 | 0.3 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.1 | 0.4 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.1 | 0.2 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.1 | 0.2 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 0.2 | GO:0003408 | optic cup formation involved in camera-type eye development(GO:0003408) |
| 0.1 | 0.2 | GO:1902617 | response to fluoride(GO:1902617) |
| 0.0 | 0.2 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.0 | 0.3 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.4 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.7 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 7.9 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 | 0.1 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 0.3 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.1 | GO:0048627 | myoblast development(GO:0048627) |
| 0.0 | 0.2 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.1 | GO:1901994 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.2 | GO:0060214 | endocardium morphogenesis(GO:0003160) endocardium formation(GO:0060214) |
| 0.0 | 0.2 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.1 | GO:0052151 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.0 | 0.2 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.1 | GO:0015766 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.3 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 0.1 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.0 | 0.1 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.2 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.0 | 0.1 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.0 | 0.1 | GO:0032900 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) negative regulation of neurotrophin production(GO:0032900) negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.1 | GO:0002276 | basophil activation involved in immune response(GO:0002276) |
| 0.0 | 0.4 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.5 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.0 | 0.1 | GO:0032484 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.0 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.1 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.1 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 0.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0042040 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.0 | 0.1 | GO:0071484 | cellular response to light intensity(GO:0071484) |
| 0.0 | 0.2 | GO:0046886 | positive regulation of hormone biosynthetic process(GO:0046886) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.0 | GO:0048371 | lateral mesodermal cell differentiation(GO:0048371) |
| 0.0 | 0.2 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.6 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.0 | 0.2 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.1 | GO:0002840 | plasmacytoid dendritic cell activation(GO:0002270) T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) regulation of restriction endodeoxyribonuclease activity(GO:0032072) |
| 0.0 | 0.5 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.0 | GO:0033092 | positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.0 | 0.2 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.3 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 0.1 | GO:0006424 | glutamyl-tRNA aminoacylation(GO:0006424) |
| 0.0 | 0.1 | GO:1903925 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.0 | 0.2 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.3 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.2 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.2 | GO:0007512 | adult heart development(GO:0007512) |
| 0.0 | 0.1 | GO:0042659 | regulation of cell fate specification(GO:0042659) |
| 0.0 | 0.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.0 | GO:1904562 | phosphatidylinositol 5-phosphate metabolic process(GO:1904562) |
| 0.0 | 0.3 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 0.0 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.1 | GO:1904046 | negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 0.0 | 0.3 | GO:2000615 | regulation of histone H3-K9 acetylation(GO:2000615) |
| 0.0 | 0.3 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 0.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.0 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.0 | 0.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.2 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.0 | 0.2 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.1 | GO:0032906 | transforming growth factor beta2 production(GO:0032906) regulation of transforming growth factor beta2 production(GO:0032909) |
| 0.0 | 0.1 | GO:1903608 | CRD-mediated mRNA stabilization(GO:0070934) protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.1 | GO:0048505 | regulation of development, heterochronic(GO:0040034) regulation of timing of cell differentiation(GO:0048505) |
| 0.0 | 0.0 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) positive regulation of reciprocal meiotic recombination(GO:0010845) negative regulation of histone phosphorylation(GO:0033128) positive regulation of meiosis I(GO:0060903) |
| 0.0 | 0.0 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.1 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.1 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.0 | GO:0021571 | rhombomere 5 development(GO:0021571) |
| 0.0 | 0.4 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) |
| 0.0 | 0.0 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.4 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.1 | GO:0051136 | extrathymic T cell differentiation(GO:0033078) regulation of NK T cell differentiation(GO:0051136) |
| 0.0 | 0.2 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.0 | 0.1 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.1 | GO:0072718 | response to cisplatin(GO:0072718) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 0.1 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.4 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.1 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.4 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.1 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.4 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.5 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.1 | GO:0031379 | RNA-directed RNA polymerase complex(GO:0031379) |
| 0.0 | 0.4 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.0 | 0.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.1 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.5 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.1 | GO:0009368 | endopeptidase Clp complex(GO:0009368) |
| 0.0 | 0.6 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.1 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.0 | 0.4 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.9 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.0 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.5 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.1 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.3 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 0.4 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.1 | 0.2 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.1 | 0.2 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.1 | 0.4 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.1 | 0.2 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.1 | 0.2 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.1 | 0.2 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.1 | 0.2 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.3 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.1 | 1.0 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 1.0 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 0.5 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.2 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.5 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.1 | GO:0004324 | ferredoxin-NADP+ reductase activity(GO:0004324) NADPH-adrenodoxin reductase activity(GO:0015039) oxidoreductase activity, acting on iron-sulfur proteins as donors(GO:0016730) oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor(GO:0016731) |
| 0.0 | 1.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.0 | 0.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.0 | 0.4 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.3 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:0015154 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.4 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 9.8 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.2 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.3 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.4 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.3 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0030305 | heparanase activity(GO:0030305) |
| 0.0 | 0.7 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.1 | GO:0061598 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.6 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.0 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.1 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0004818 | glutamate-tRNA ligase activity(GO:0004818) |
| 0.0 | 1.6 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.3 | GO:0033549 | MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.3 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.0 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.2 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.2 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.2 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.2 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.0 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 0.5 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.1 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 1.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.3 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.5 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.5 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.4 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.9 | PID TNF PATHWAY | TNF receptor signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.4 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.6 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.4 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.8 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 1.0 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.3 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 1.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.2 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.4 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |