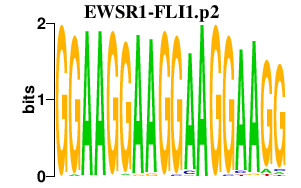
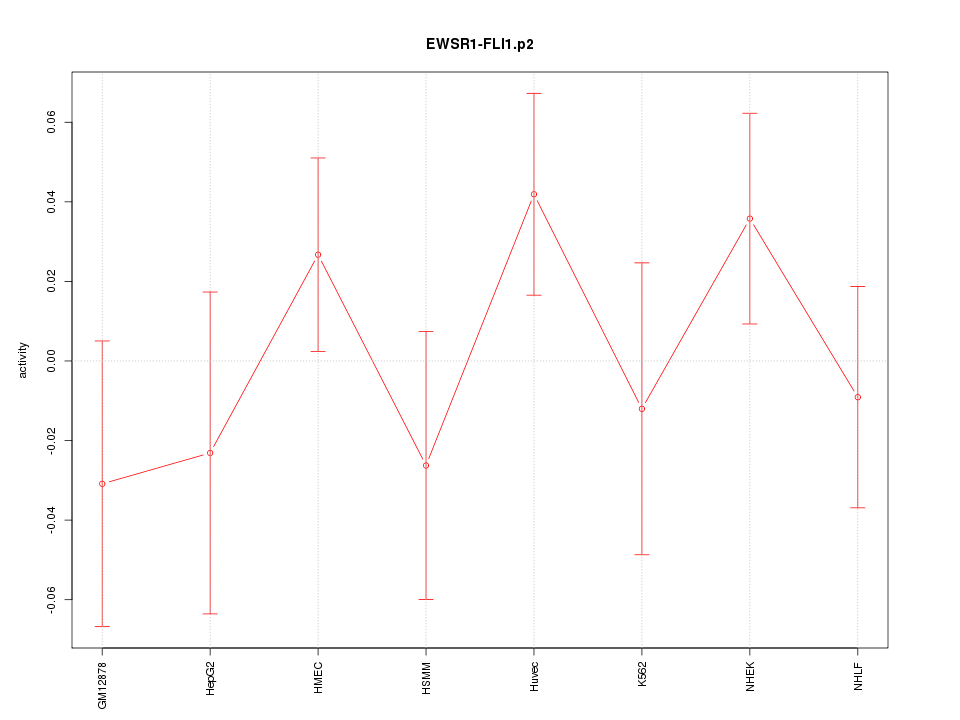
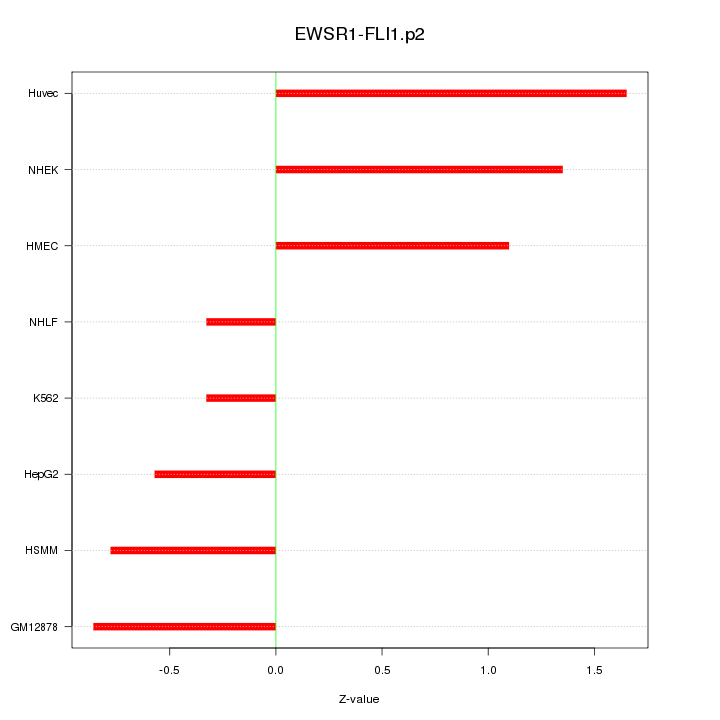
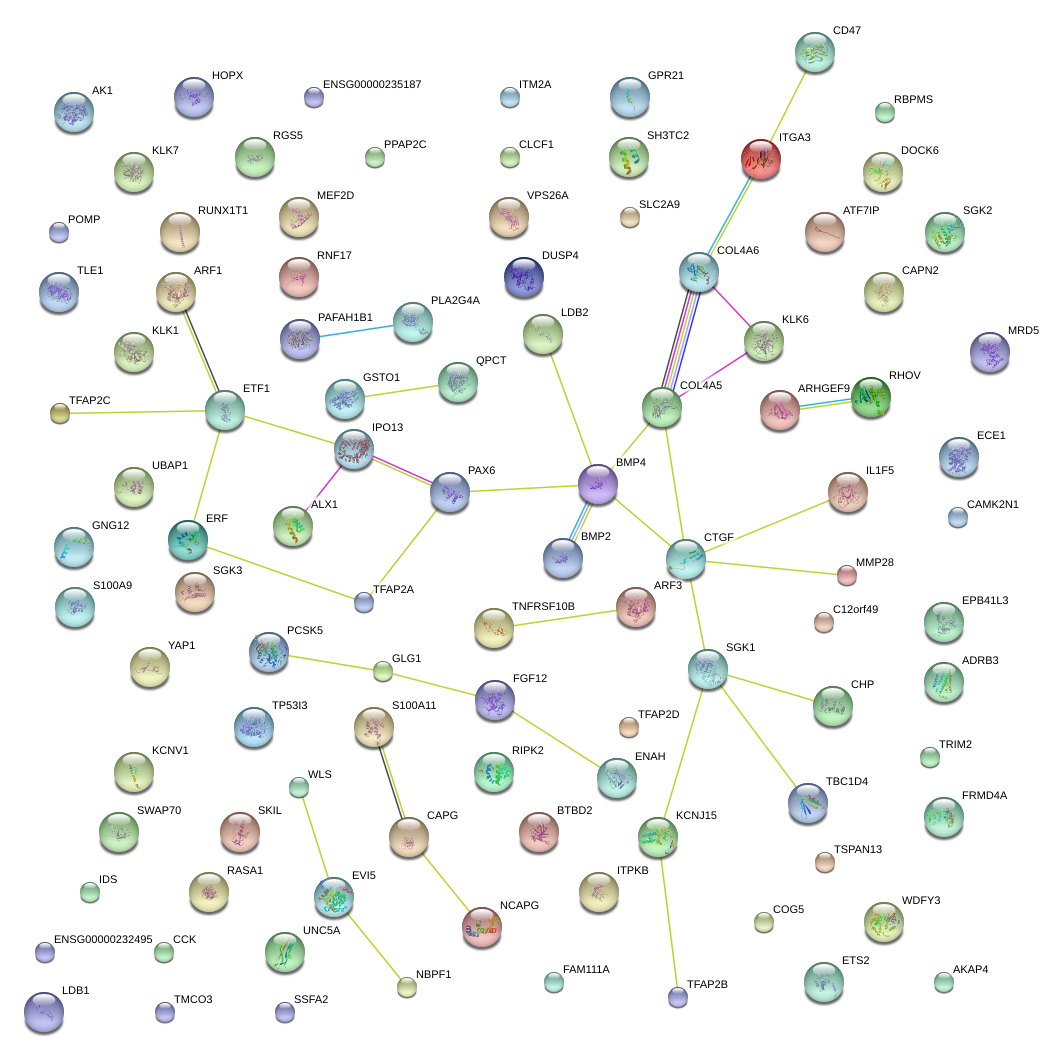
|
chr1_-_68071635
|
1.926
|
NM_018841
|
GNG12
|
guanine nucleotide binding protein (G protein), gamma 12
|
|
chr7_+_16759845
|
1.875
|
NM_014399
|
TSPAN13
|
tetraspanin 13
|
|
chr6_-_10520592
|
1.668
|
NM_001032280
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha)
|
|
chr1_+_185064654
|
1.490
|
NM_024420
|
PLA2G4A
|
phospholipase A2, group IVA (cytosolic, calcium-dependent)
|
|
chr1_-_150276064
|
1.465
|
|
S100A11
|
S100 calcium binding protein A11
|
|
chr1_-_150276091
|
1.415
|
|
S100A11
|
S100 calcium binding protein A11
|
|
chr1_-_68470585
|
1.265
|
NM_001002292
NM_001193334
NM_024911
|
WLS
|
wntless homolog (Drosophila)
|
|
chr1_+_221955876
|
1.225
|
NM_001146068
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chrX_+_107569671
|
1.168
|
NM_000495
NM_033380
|
COL4A5
|
collagen, type IV, alpha 5
|
|
chr6_-_134538762
|
1.093
|
NM_001143678
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr6_-_134538526
|
1.066
|
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr3_-_42282665
|
0.992
|
NM_000729
|
CCK
|
cholecystokinin
|
|
chr1_-_150276114
|
0.966
|
NM_005620
|
S100A11
|
S100 calcium binding protein A11
|
|
chr2_+_37425240
|
0.919
|
NM_012413
|
QPCT
|
glutaminyl-peptide cyclotransferase
|
|
chr2_-_24161033
|
0.847
|
NM_147184
NM_004881
|
TP53I3
|
tumor protein p53 inducible protein 3
|
|
chr6_-_132314004
|
0.750
|
NM_001901
|
CTGF
|
connective tissue growth factor
|
|
chr17_+_45488330
|
0.649
|
NM_002204
NM_005501
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor)
|
|
chr17_+_45488356
|
0.639
|
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor)
|
|
chr2_+_182464910
|
0.612
|
|
SSFA2
|
sperm specific antigen 2
|
|
chr17_+_2443983
|
0.604
|
|
PAFAH1B1
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 (45kDa)
|
|
chr1_-_93030538
|
0.526
|
NM_005665
|
EVI5
|
ecotropic viral integration site 5
|
|
chr1_-_223907283
|
0.517
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr17_+_2443652
|
0.512
|
NM_000430
|
PAFAH1B1
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 (45kDa)
|
|
chrX_-_107569256
|
0.504
|
NM_001847
|
COL4A6
|
collagen, type IV, alpha 6
|
|
chr18_-_5620629
|
0.480
|
|
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3
|
|
chr20_+_6696744
|
0.473
|
NM_001200
|
BMP2
|
bone morphogenetic protein 2
|
|
chr11_+_101485946
|
0.462
|
NM_001130145
NM_001195044
NM_006106
|
YAP1
|
Yes-associated protein 1
|
|
chr8_-_22982435
|
0.429
|
NM_003842
NM_147187
|
TNFRSF10B
|
tumor necrosis factor receptor superfamily, member 10b
|
|
chrX_-_148394570
|
0.414
|
NM_000202
NM_001166550
NM_006123
|
IDS
|
iduronate 2-sulfatase
|
|
chr19_-_242168
|
0.405
|
NM_177543
|
PPAP2C
|
phosphatidic acid phosphatase type 2C
|
|
chr12_-_47637530
|
0.382
|
|
|
|
|
chr5_-_148422799
|
0.376
|
NM_024577
|
SH3TC2
|
SH3 domain and tetratricopeptide repeats 2
|
|
chr4_-_16509354
|
0.371
|
|
LDB2
|
LIM domain binding 2
|
|
chr4_-_86106567
|
0.366
|
NM_014991
|
WDFY3
|
WD repeat and FYVE domain containing 3
|
|
chr12_-_115660017
|
0.363
|
NM_024738
|
C12orf49
|
chromosome 12 open reading frame 49
|
|
chr4_-_16509446
|
0.362
|
NM_001130834
NM_001290
|
LDB2
|
LIM domain binding 2
|
|
chr4_-_16509126
|
0.328
|
|
LDB2
|
LIM domain binding 2
|
|
chr2_+_113532685
|
0.309
|
NM_173170
|
IL1F5
|
interleukin 1 family, member 5 (delta)
|
|
chr9_-_83494198
|
0.296
|
|
TLE1
|
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila)
|
|
chr12_-_47637396
|
0.287
|
|
ARF3
|
ADP-ribosylation factor 3
|
|
chr21_+_38550533
|
0.285
|
NM_002243
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15
|
|
chr17_-_31146660
|
0.281
|
NM_001032278
NM_024302
|
MMP28
|
matrix metallopeptidase 28
|
|
chr12_-_47637507
|
0.280
|
|
ARF3
|
ADP-ribosylation factor 3
|
|
chr10_+_70553983
|
0.263
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr12_-_47637564
|
0.255
|
|
ARF3
|
ADP-ribosylation factor 3
|
|
chr4_-_57216816
|
0.255
|
|
HOPX
|
HOP homeobox
|
|
chr19_-_1966628
|
0.254
|
NM_017797
|
BTBD2
|
BTB (POZ) domain containing 2
|
|
chr19_-_242335
|
0.253
|
NM_003712
NM_177526
|
PPAP2C
|
phosphatidic acid phosphatase type 2C
|
|
chr12_+_84198015
|
0.247
|
NM_006982
|
ALX1
|
ALX homeobox 1
|
|
chr12_-_47637512
|
0.245
|
|
ARF3
|
ADP-ribosylation factor 3
|
|
chr12_-_47637518
|
0.245
|
NM_001659
|
ARF3
|
ADP-ribosylation factor 3
|
|
chr2_+_182464713
|
0.244
|
NM_001130445
NM_006751
|
SSFA2
|
sperm specific antigen 2
|
|
chr1_-_20685314
|
0.243
|
NM_018584
|
CAMK2N1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr1_-_224993498
|
0.236
|
NM_002221
|
ITPKB
|
inositol 1,4,5-trisphosphate 3-kinase B
|
|
chr7_-_106991941
|
0.229
|
NM_001161520
NM_006348
NM_181733
|
COG5
|
component of oligomeric golgi complex 5
|
|
chr1_+_151596953
|
0.226
|
NM_002965
|
S100A9
|
S100 calcium binding protein A9
|
|
chr21_+_38550739
|
0.225
|
NM_170736
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15
|
|
chr10_+_70553993
|
0.223
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr5_+_86599350
|
0.222
|
|
RASA1
|
RAS p21 protein activator (GTPase activating protein) 1
|
|
chr8_+_30361485
|
0.214
|
NM_001008710
NM_001008711
NM_001008712
NM_006867
|
RBPMS
|
RNA binding protein with multiple splicing
|
|
chr4_+_154345039
|
0.204
|
NM_015271
|
TRIM2
|
tripartite motif containing 2
|
|
chr4_-_57217226
|
0.202
|
NM_001145459
NM_139211
|
HOPX
|
HOP homeobox
|
|
chr12_+_14409875
|
0.201
|
NM_018179
|
ATF7IP
|
activating transcription factor 7 interacting protein
|
|
chr14_-_53493198
|
0.200
|
NM_001202
NM_130850
|
BMP4
|
bone morphogenetic protein 4
|
|
chr8_-_93177057
|
0.198
|
NM_001198625
NM_001198626
NM_001198627
NM_001198631
NM_001198634
NM_001198679
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr9_+_34169023
|
0.197
|
|
UBAP1
|
ubiquitin associated protein 1
|
|
chr10_-_103864651
|
0.187
|
NM_003893
|
LDB1
|
LIM domain binding 1
|
|
chr9_+_34169036
|
0.186
|
|
UBAP1
|
ubiquitin associated protein 1
|
|
chr9_-_129679760
|
0.184
|
NM_000476
|
AK1
|
adenylate kinase 1
|
|
chr10_+_106004457
|
0.175
|
NM_001191002
NM_004832
|
GSTO1
|
glutathione S-transferase omega 1
|
|
chr11_-_66897584
|
0.174
|
NM_013246
|
CLCF1
|
cardiotrophin-like cytokine factor 1
|
|
chr10_+_70553936
|
0.169
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chrX_-_78509662
|
0.163
|
NM_001171581
NM_004867
|
ITM2A
|
integral membrane protein 2A
|
|
chr1_+_44185005
|
0.158
|
NM_014652
|
IPO13
|
importin 13
|
|
chr13_-_74953873
|
0.157
|
NM_014832
|
TBC1D4
|
TBC1 domain family, member 4
|
|
chr10_+_70553862
|
0.156
|
NM_001035260
NM_004896
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr1_-_154737189
|
0.154
|
|
MEF2D
|
myocyte enhancer factor 2D
|
|
chr8_-_29262240
|
0.149
|
NM_057158
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr13_+_28131279
|
0.146
|
|
POMP
|
proteasome maturation protein
|
|
chr19_-_56178882
|
0.141
|
NM_139277
NM_005046
|
KLK7
|
kallikrein-related peptidase 7
|
|
chr19_-_47450991
|
0.141
|
|
ERF
|
Ets2 repressor factor
|
|
chrX_-_78509367
|
0.139
|
|
ITM2A
|
integral membrane protein 2A
|
|
chr16_-_73198515
|
0.136
|
NM_001145666
NM_001145667
NM_012201
|
GLG1
|
golgi glycoprotein 1
|
|
chr9_+_34169002
|
0.135
|
NM_001171201
NM_001171202
NM_001171203
NM_001171204
NM_016525
|
UBAP1
|
ubiquitin associated protein 1
|
|
chr10_-_14412813
|
0.133
|
NM_018027
|
FRMD4A
|
FERM domain containing 4A
|
|
chr2_+_113533155
|
0.133
|
NM_012275
|
IL1F5
|
interleukin 1 family, member 5 (delta)
|
|
chr19_-_56164628
|
0.132
|
NM_002774
|
KLK6
|
kallikrein-related peptidase 6
|
|
chr5_+_86599800
|
0.131
|
|
RASA1
|
RAS p21 protein activator (GTPase activating protein) 1
|
|
chr9_+_34169058
|
0.131
|
|
UBAP1
|
ubiquitin associated protein 1
|
|
chr11_+_58668827
|
0.129
|
NM_022074
NM_198847
|
FAM111A
|
family with sequence similarity 111, member A
|
|
chrX_-_49851743
|
0.129
|
NM_139289
|
AKAP4
|
A kinase (PRKA) anchor protein 4
|
|
chr21_+_39099717
|
0.128
|
NM_005239
|
ETS2
|
v-ets erythroblastosis virus E26 oncogene homolog 2 (avian)
|
|
chr8_-_37943143
|
0.127
|
NM_000025
|
ADRB3
|
adrenergic, beta-3-, receptor
|
|
chr13_+_113193308
|
0.127
|
NM_017905
|
TMCO3
|
transmembrane and coiled-coil domains 3
|
|
chr11_+_9642226
|
0.127
|
|
SWAP70
|
SWAP switching B-cell complex 70kDa subunit
|
|
chr13_+_28131140
|
0.126
|
NM_015932
|
POMP
|
proteasome maturation protein
|
|
chr9_+_124836666
|
0.125
|
NM_005294
|
GPR21
|
G protein-coupled receptor 21
|
|
chr20_+_41621119
|
0.125
|
|
SGK2
|
serum/glucocorticoid regulated kinase 2
|
|
chr10_-_103869923
|
0.123
|
NM_001113407
|
LDB1
|
LIM domain binding 1
|
|
chr11_-_31789168
|
0.123
|
|
PAX6
|
paired box 6
|
|
chr19_-_47451119
|
0.123
|
NM_006494
|
ERF
|
Ets2 repressor factor
|
|
chr15_+_39310693
|
0.120
|
NM_007236
|
CHP
|
calcium binding protein P22
|
|
chr15_+_39310757
|
0.114
|
|
CHP
|
calcium binding protein P22
|
|
chr3_-_109292444
|
0.114
|
NM_001777
NM_198793
|
CD47
|
CD47 molecule
|
|
chr8_-_93176819
|
0.111
|
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr3_+_171558143
|
0.110
|
NM_001145098
NM_005414
|
SKIL
|
SKI-like oncogene
|
|
chrX_-_62891712
|
0.109
|
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9
|
|
chr1_-_161379606
|
0.108
|
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr8_-_111056099
|
0.101
|
NM_014379
|
KCNV1
|
potassium channel, subfamily V, member 1
|
|
chr10_+_70553949
|
0.100
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr9_+_77695334
|
0.093
|
NM_001190482
NM_006200
|
PCSK5
|
proprotein convertase subtilisin/kexin type 5
|
|
chr19_-_11234117
|
0.092
|
NM_020812
|
DOCK6
|
dedicator of cytokinesis 6
|
|
chr1_-_21478571
|
0.092
|
NM_001113347
|
ECE1
|
endothelin converting enzyme 1
|
|
chr3_+_171558188
|
0.089
|
|
SKIL
|
SKI-like oncogene
|
|
chr3_-_193928038
|
0.088
|
NM_004113
|
FGF12
|
fibroblast growth factor 12
|
|
chr2_-_85490997
|
0.087
|
NM_001747
|
CAPG
|
capping protein (actin filament), gelsolin-like
|
|
chr4_-_9650931
|
0.086
|
NM_001001290
|
SLC2A9
|
solute carrier family 2 (facilitated glucose transporter), member 9
|
|
chr16_-_73198416
|
0.086
|
|
GLG1
|
golgi glycoprotein 1
|
|
chr12_+_29193399
|
0.085
|
|
FAR2
|
fatty acyl CoA reductase 2
|
|
chr9_+_35528890
|
0.084
|
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chrX_+_67635593
|
0.082
|
|
YIPF6
|
Yip1 domain family, member 6
|
|
chr14_+_22845851
|
0.079
|
NM_004050
|
BCL2L2-PABPN1
BCL2L2
|
BCL2L2-PABPN1 read-through transcript
BCL2-like 2
|
|
chr1_+_109036431
|
0.078
|
NM_018061
|
PRPF38B
|
PRP38 pre-mRNA processing factor 38 (yeast) domain containing B
|
|
chr10_+_23424383
|
0.077
|
NM_012228
|
MSRB2
|
methionine sulfoxide reductase B2
|
|
chr8_-_93176618
|
0.077
|
NM_001198629
NM_001198630
NM_001198632
NM_175635
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr11_-_62231358
|
0.073
|
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin)
|
|
chr10_-_35143900
|
0.073
|
NM_001184785
NM_001184786
NM_001184787
NM_001184788
NM_001184789
NM_001184790
NM_001184791
NM_001184792
NM_001184793
NM_001184794
NM_019619
|
PARD3
|
par-3 partitioning defective 3 homolog (C. elegans)
|
|
chr9_-_83493415
|
0.071
|
NM_005077
|
TLE1
|
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila)
|
|
chr8_-_145530639
|
0.071
|
NM_031309
|
SCRT1
|
scratch homolog 1, zinc finger protein (Drosophila)
|
|
chr4_+_174326464
|
0.071
|
NM_017423
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7)
|
|
chrY_+_1361570
|
0.071
|
NM_001161530
NM_172247
|
CSF2RA
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage)
|
|
chrX_+_1361570
|
0.071
|
NM_001161530
NM_172247
|
CSF2RA
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage)
|
|
chr20_+_29656744
|
0.070
|
NM_002165
NM_181353
|
ID1
|
inhibitor of DNA binding 1, dominant negative helix-loop-helix protein
|
|
chr9_-_69669550
|
0.070
|
NM_001099279
NM_199244
|
FOXD4L2
FOXD4L4
|
forkhead box D4-like 2
forkhead box D4-like 4
|
|
chr1_+_152460381
|
0.069
|
|
UBAP2L
|
ubiquitin associated protein 2-like
|
|
chr12_+_54838408
|
0.067
|
|
MYL6
|
myosin, light chain 6, alkali, smooth muscle and non-muscle
|
|
chr19_-_52243721
|
0.066
|
NM_017854
|
TMEM160
|
transmembrane protein 160
|
|
chr5_+_175041069
|
0.065
|
NM_022304
|
HRH2
|
histamine receptor H2
|
|
chr3_+_14141583
|
0.063
|
|
TMEM43
|
transmembrane protein 43
|
|
chr17_+_35372751
|
0.062
|
NM_178171
|
GSDMA
|
gasdermin A
|
|
chr10_-_102017327
|
0.062
|
NM_018294
|
CWF19L1
|
CWF19-like 1, cell cycle control (S. pombe)
|
|
chr11_+_2279812
|
0.062
|
NM_139022
|
TSPAN32
|
tetraspanin 32
|
|
chr16_-_73198498
|
0.062
|
|
GLG1
|
golgi glycoprotein 1
|
|
chr1_-_231498081
|
0.061
|
NM_014801
|
PCNXL2
|
pecanex-like 2 (Drosophila)
|
|
chr15_-_71712605
|
0.061
|
|
NPTN
|
neuroplastin
|
|
chr12_+_54838733
|
0.060
|
|
MYL6
|
myosin, light chain 6, alkali, smooth muscle and non-muscle
|
|
chr1_+_153241665
|
0.058
|
NM_015872
|
ZBTB7B
|
zinc finger and BTB domain containing 7B
|
|
chr12_+_121803255
|
0.057
|
NM_003677
|
DENR
|
density-regulated protein
|
|
chr17_+_4784524
|
0.057
|
|
RNF167
|
ring finger protein 167
|
|
chr3_-_129852345
|
0.057
|
|
RPN1
|
ribophorin I
|
|
chrX_-_62891667
|
0.055
|
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9
|
|
chr1_+_152459918
|
0.054
|
NM_014847
|
UBAP2L
|
ubiquitin associated protein 2-like
|
|
chr12_+_77781903
|
0.052
|
NM_001135805
|
SYT1
|
synaptotagmin I
|
|
chr8_+_22078622
|
0.052
|
|
BMP1
|
bone morphogenetic protein 1
|
|
chr12_+_47945132
|
0.052
|
|
TUBA1C
|
tubulin, alpha 1c
|
|
chr11_-_8789457
|
0.052
|
NM_139157
|
ST5
|
suppression of tumorigenicity 5
|
|
chr12_-_9651748
|
0.051
|
NM_002258
|
KLRB1
|
killer cell lectin-like receptor subfamily B, member 1
|
|
chr11_+_113435506
|
0.050
|
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr8_-_27751462
|
0.049
|
|
PBK
|
PDZ binding kinase
|
|
chr11_-_62231386
|
0.049
|
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin)
|
|
chr16_-_74126568
|
0.047
|
NM_024533
|
CHST5
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 5
|
|
chr6_-_46091238
|
0.047
|
NM_016929
|
CLIC5
|
chloride intracellular channel 5
|
|
chr3_-_121295663
|
0.045
|
NM_001146156
NM_002093
|
GSK3B
|
glycogen synthase kinase 3 beta
|
|
chr3_-_129852400
|
0.044
|
NM_002950
|
RPN1
|
ribophorin I
|
|
chr17_+_53625084
|
0.043
|
NM_000502
|
EPX
|
eosinophil peroxidase
|
|
chr11_-_62231636
|
0.042
|
NM_001122955
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin)
|
|
chr11_+_17714070
|
0.041
|
NM_001112741
NM_004976
|
KCNC1
|
potassium voltage-gated channel, Shaw-related subfamily, member 1
|
|
chr12_+_47945086
|
0.041
|
NM_032704
|
TUBA1C
|
tubulin, alpha 1c
|
|
chr3_+_33130451
|
0.041
|
NM_006371
|
CRTAP
|
cartilage associated protein
|
|
chr1_-_221604023
|
0.040
|
NM_001037175
NM_017982
|
SUSD4
|
sushi domain containing 4
|
|
chr5_-_64100176
|
0.040
|
|
SREK1IP1
|
SREK1-interacting protein 1
|
|
chrX_+_80344253
|
0.040
|
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like
|
|
chr1_+_153241858
|
0.039
|
|
ZBTB7B
|
zinc finger and BTB domain containing 7B
|
|
chr14_+_55654607
|
0.039
|
NM_021255
|
PELI2
|
pellino homolog 2 (Drosophila)
|
|
chr11_-_62231445
|
0.038
|
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin)
|
|
chr2_-_222867171
|
0.037
|
|
PAX3
|
paired box 3
|
|
chrX_-_62891730
|
0.037
|
NM_001173480
NM_015185
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9
|
|
chr9_+_35528628
|
0.036
|
NM_001135999
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr12_+_107797985
|
0.035
|
NM_001917
|
DAO
|
D-amino-acid oxidase
|
|
chr7_+_65178369
|
0.035
|
|
ASL
|
argininosuccinate lyase
|
|
chr11_-_66898223
|
0.035
|
NM_001166212
|
CLCF1
|
cardiotrophin-like cytokine factor 1
|
|
chr5_-_64100239
|
0.034
|
NM_173829
|
SREK1IP1
|
SREK1-interacting protein 1
|
|
chr11_-_8788765
|
0.034
|
NM_213618
|
ST5
|
suppression of tumorigenicity 5
|
|
chr19_-_11355882
|
0.033
|
NM_000121
|
EPOR
|
erythropoietin receptor
|
|
chr18_-_44189625
|
0.033
|
|
ZBTB7C
|
zinc finger and BTB domain containing 7C
|
|
chr8_+_22078589
|
0.032
|
NM_001199
NM_006129
|
BMP1
|
bone morphogenetic protein 1
|
|
chrX_+_2994852
|
0.032
|
NM_004042
|
ARSF
|
arylsulfatase F
|
|
chr3_-_50353209
|
0.031
|
NM_007182
NM_170714
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1
|
|
chr14_+_55654900
|
0.030
|
|
PELI2
|
pellino homolog 2 (Drosophila)
|
|
chr11_+_2279932
|
0.030
|
|
TSPAN32
|
tetraspanin 32
|
|
chr19_+_57592913
|
0.029
|
NM_032423
|
ZNF528
|
zinc finger protein 528
|
|
chr11_-_62231242
|
0.029
|
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin)
|
|
chr3_+_33130563
|
0.029
|
|
CRTAP
|
cartilage associated protein
|
|
chr7_-_138133388
|
0.029
|
NM_020632
NM_130840
|
ATP6V0A4
|
ATPase, H+ transporting, lysosomal V0 subunit a4
|
|
chr10_+_114699953
|
0.028
|
NM_001146274
NM_001146283
NM_001146284
NM_001146285
NM_001146286
NM_001198525
NM_001198526
NM_001198527
NM_001198528
NM_001198529
NM_001198530
NM_001198531
NM_030756
|
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box)
|
|
chr14_-_20636568
|
0.028
|
NM_016423
|
ZNF219
|
zinc finger protein 219
|
|
chr11_-_62231565
|
0.027
|
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin)
|
|
chr11_-_62213946
|
0.027
|
NM_203422
|
LRRN4CL
|
LRRN4 C-terminal like
|
|
chrX_+_56606686
|
0.026
|
NM_013444
|
UBQLN2
|
ubiquilin 2
|
|
chr1_-_148166247
|
0.026
|
|
SF3B4
|
splicing factor 3b, subunit 4, 49kDa
|