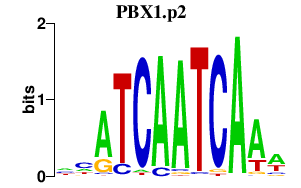
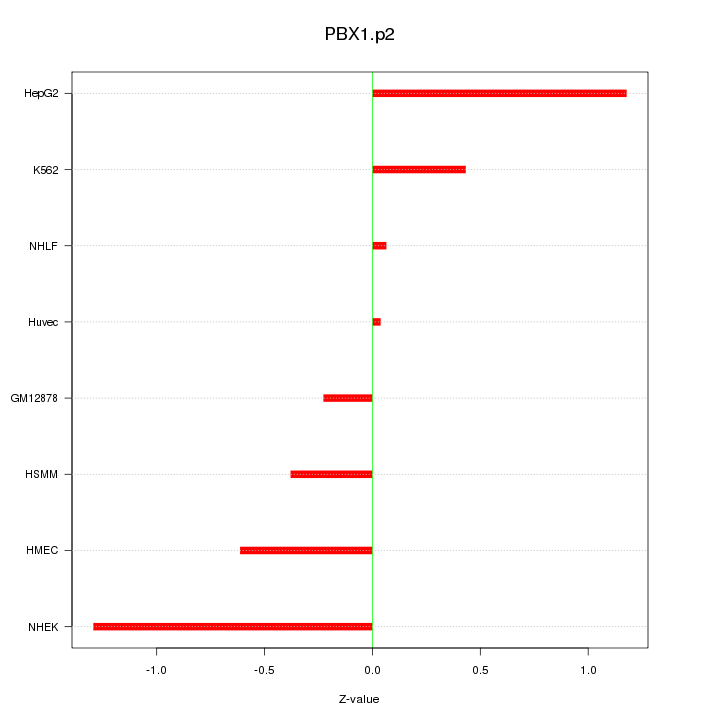
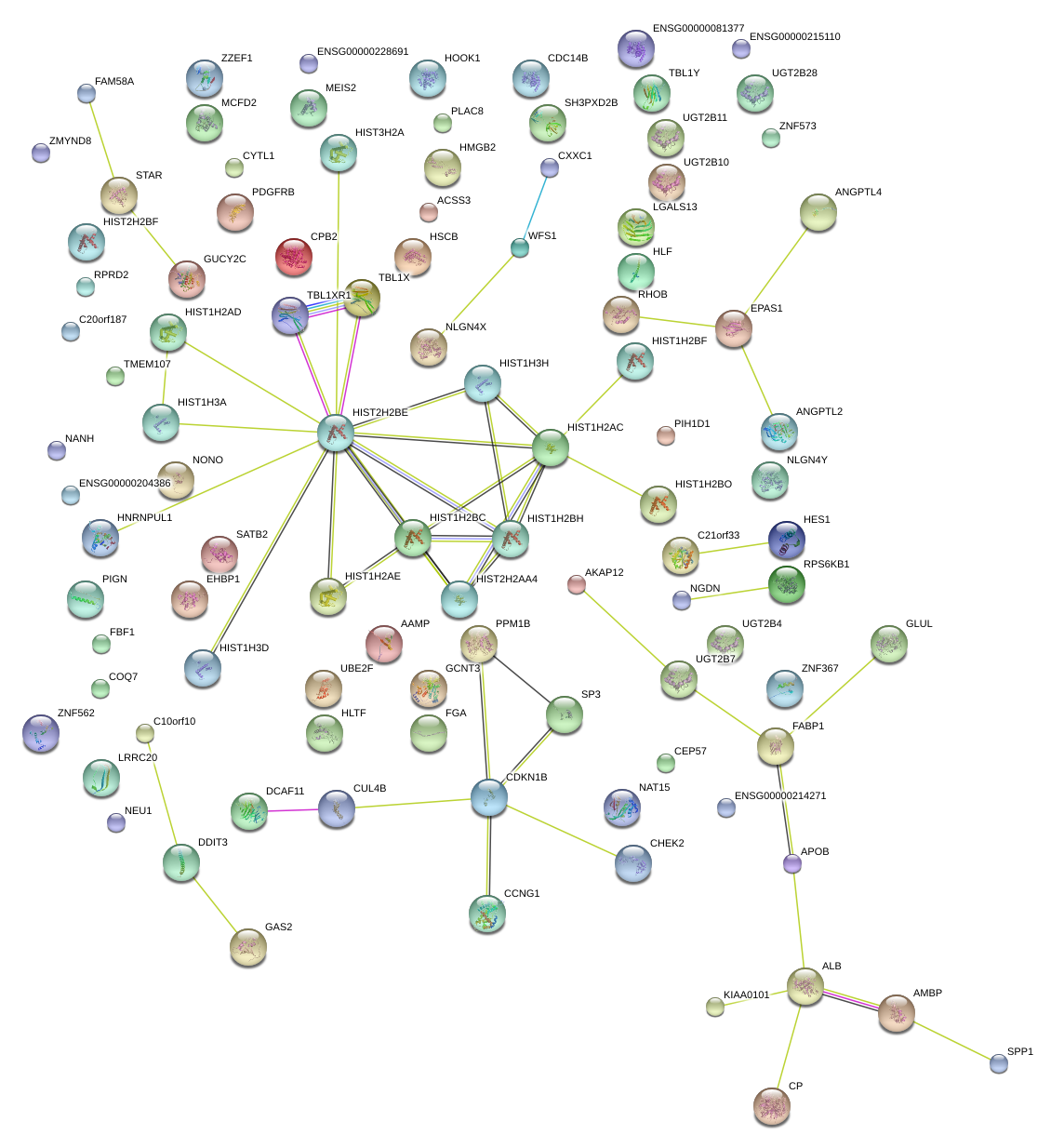
|
chr13_-_45577144
|
1.314
|
NM_001872
NM_016413
|
CPB2
|
carboxypeptidase B2 (plasma)
|
|
chr4_+_74496575
|
0.859
|
|
ALB
|
albumin
|
|
chr17_+_16225130
|
0.829
|
|
|
|
|
chr4_+_74499651
|
0.829
|
|
ALB
|
albumin
|
|
chr22_-_27467776
|
0.763
|
NM_001005735
NM_007194
NM_145862
|
CHEK2
|
CHK2 checkpoint homolog (S. pombe)
|
|
chr9_-_115880366
|
0.755
|
|
AMBP
|
alpha-1-microglobulin/bikunin precursor
|
|
chr9_-_115880281
|
0.746
|
|
AMBP
|
alpha-1-microglobulin/bikunin precursor
|
|
chr9_-_115880425
|
0.716
|
|
AMBP
|
alpha-1-microglobulin/bikunin precursor
|
|
chr14_+_23654161
|
0.683
|
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr2_+_20510312
|
0.614
|
NM_004040
|
RHOB
|
ras homolog gene family, member B
|
|
chr4_+_74496660
|
0.591
|
|
ALB
|
albumin
|
|
chr20_-_45410033
|
0.578
|
|
ZMYND8
|
zinc finger, MYND-type containing 8
|
|
chr2_-_200030944
|
0.531
|
NM_001172509
|
SATB2
|
SATB homeobox 2
|
|
chr5_+_162797149
|
0.517
|
NM_004060
NM_199246
|
CCNG1
|
cyclin G1
|
|
chr4_+_69716298
|
0.483
|
NM_001075
NM_001144767
|
UGT2B10
|
UDP glucuronosyltransferase 2 family, polypeptide B10
|
|
chr15_-_35179185
|
0.467
|
|
MEIS2
|
Meis homeobox 2
|
|
chr9_-_98421894
|
0.440
|
NM_003671
NM_033331
|
CDC14B
|
CDC14 cell division cycle 14 homolog B (S. cerevisiae)
|
|
chr15_-_35178888
|
0.440
|
NM_172316
|
MEIS2
|
Meis homeobox 2
|
|
chr15_-_35180656
|
0.421
|
NM_002399
|
MEIS2
|
Meis homeobox 2
|
|
chr1_+_15862887
|
0.419
|
|
|
|
|
chr9_-_115880372
|
0.412
|
|
AMBP
|
alpha-1-microglobulin/bikunin precursor
|
|
chr14_+_23653848
|
0.404
|
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr14_+_23653819
|
0.398
|
NM_025230
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr2_-_174538197
|
0.397
|
|
SP3
|
Sp3 transcription factor
|
|
chr14_+_23008723
|
0.376
|
NM_001042635
NM_015514
|
NGDN
|
neuroguidin, EIF4E binding protein
|
|
chr12_+_79995927
|
0.376
|
NM_024560
|
ACSS3
|
acyl-CoA synthetase short-chain family member 3
|
|
chr11_+_95163256
|
0.374
|
NM_014679
|
CEP57
|
centrosomal protein 57kDa
|
|
chr4_-_174492029
|
0.366
|
NM_001130688
NM_002129
|
HMGB2
|
high-mobility group box 2
|
|
chr2_-_88208650
|
0.365
|
NM_001443
|
FABP1
|
fatty acid binding protein 1, liver
|
|
chr1_-_180627512
|
0.360
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chrX_+_9391314
|
0.350
|
NM_001139466
NM_001139467
NM_001139468
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr6_-_31938572
|
0.350
|
NM_000434
|
NEU1
|
sialidase 1 (lysosomal sialidase)
|
|
chr2_-_21079145
|
0.349
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr14_+_23653745
|
0.346
|
NM_001163484
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr9_-_115880494
|
0.342
|
NM_001633
|
AMBP
|
alpha-1-microglobulin/bikunin precursor
|
|
chr12_+_12761085
|
0.333
|
NM_004064
|
CDKN1B
|
cyclin-dependent kinase inhibitor 1B (p27, Kip1)
|
|
chr11_+_95163457
|
0.326
|
|
CEP57
|
centrosomal protein 57kDa
|
|
chr3_+_195336631
|
0.307
|
|
HES1
|
hairy and enhancer of split 1, (Drosophila)
|
|
chr14_+_23008742
|
0.306
|
|
NGDN
|
neuroguidin, EIF4E binding protein
|
|
chr11_+_22646229
|
0.305
|
NM_177553
|
GAS2
|
growth arrest-specific 2
|
|
chr6_+_26307691
|
0.305
|
NM_003522
|
HIST1H2BF
|
histone cluster 1, H2bf
|
|
chr4_-_70115037
|
0.286
|
NM_001073
|
UGT2B11
|
UDP glucuronosyltransferase 2 family, polypeptide B11
|
|
chr3_+_195336625
|
0.285
|
NM_005524
|
HES1
|
hairy and enhancer of split 1, (Drosophila)
|
|
chr3_-_150286812
|
0.271
|
NM_003071
NM_139048
|
HLTF
|
helicase-like transcription factor
|
|
chr6_-_31938530
|
0.267
|
|
NEU1
|
sialidase 1 (lysosomal sialidase)
|
|
chr16_+_18986459
|
0.267
|
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chr16_+_2593351
|
0.260
|
|
LOC652276
|
hypothetical LOC652276
|
|
chrY_+_15145019
|
0.260
|
NM_014893
|
NLGN4Y
|
neuroligin 4, Y-linked
|
|
chr4_+_70180805
|
0.258
|
NM_053039
|
UGT2B28
|
UDP glucuronosyltransferase 2 family, polypeptide B28
|
|
chr5_-_171814026
|
0.256
|
NM_001017995
|
SH3PXD2B
|
SH3 and PX domains 2B
|
|
chr19_-_54646837
|
0.255
|
NM_017916
|
PIH1D1
|
PIH1 domain containing 1
|
|
chr4_+_89115892
|
0.247
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr2_+_238540361
|
0.243
|
|
UBE2F
|
ubiquitin-conjugating enzyme E2F (putative)
|
|
chr2_+_44249487
|
0.241
|
NM_001033556
NM_001033557
NM_002706
NM_177968
NM_177969
|
PPM1B
|
protein phosphatase, Mg2+/Mn2+ dependent, 1B
|
|
chr16_+_18986417
|
0.240
|
NM_016138
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chr4_+_6322459
|
0.239
|
NM_001145853
NM_006005
|
WFS1
|
Wolfram syndrome 1 (wolframin)
|
|
chr5_-_149515613
|
0.232
|
NM_002609
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr4_-_155731288
|
0.226
|
NM_000508
NM_021871
|
FGA
|
fibrinogen alpha chain
|
|
chr1_+_60053050
|
0.225
|
NM_015888
|
HOOK1
|
hook homolog 1 (Drosophila)
|
|
chr4_+_89115862
|
0.224
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr3_-_150422474
|
0.224
|
NM_000096
|
CP
|
ceruloplasmin (ferroxidase)
|
|
chr10_-_71812350
|
0.219
|
NM_018205
NM_018239
|
LRRC20
|
leucine rich repeat containing 20
|
|
chr16_+_3433606
|
0.214
|
NM_001083601
|
NAT15
|
N-acetyltransferase 15 (GCN5-related, putative)
|
|
chr6_+_151603201
|
0.214
|
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr15_+_57696016
|
0.214
|
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type
|
|
chr6_+_26232351
|
0.212
|
NM_003512
|
HIST1H2AC
|
histone cluster 1, H2ac
|
|
chr9_-_128910132
|
0.209
|
|
ANGPTL2
|
angiopoietin-like 2
|
|
chr4_+_89115825
|
0.204
|
NM_000582
NM_001040058
NM_001040060
|
SPP1
|
secreted phosphoprotein 1
|
|
chr2_-_46988449
|
0.202
|
|
MCFD2
|
multiple coagulation factor deficiency 2
|
|
chr18_-_58005268
|
0.200
|
NM_012327
NM_176787
|
PIGN
|
phosphatidylinositol glycan anchor biosynthesis, class N
|
|
chr22_+_27468018
|
0.200
|
NM_172002
|
HSCB
|
HscB iron-sulfur cluster co-chaperone homolog (E. coli)
|
|
chr17_+_50697319
|
0.198
|
NM_002126
|
HLF
|
hepatic leukemia factor
|
|
chr12_-_56200547
|
0.195
|
NM_001195053
NM_001195054
NM_001195055
NM_001195056
NM_001195057
NM_004083
|
DDIT3
|
DNA-damage-inducible transcript 3
|
|
chr4_+_6322489
|
0.195
|
|
WFS1
|
Wolfram syndrome 1 (wolframin)
|
|
chr4_-_84254912
|
0.195
|
|
PLAC8
|
placenta-specific 8
|
|
chr8_-_38127489
|
0.194
|
NM_000349
|
STAR
|
steroidogenic acute regulatory protein
|
|
chr19_+_46462113
|
0.193
|
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1
|
|
chr15_-_62460622
|
0.192
|
NM_001029989
NM_014736
|
KIAA0101
|
KIAA0101
|
|
chr2_+_62786290
|
0.189
|
NM_001142616
NM_015252
|
EHBP1
|
EH domain binding protein 1
|
|
chr2_-_218843039
|
0.188
|
|
AAMP
|
angio-associated, migratory cell protein
|
|
chr2_-_218843109
|
0.180
|
NM_001087
|
AAMP
|
angio-associated, migratory cell protein
|
|
chr6_-_26307418
|
0.178
|
NM_021065
NM_003530
|
HIST1H2AD
HIST1H3D
|
histone cluster 1, H2ad
histone cluster 1, H3d
|
|
chrX_-_119550402
|
0.178
|
|
CUL4B
|
cullin 4B
|
|
chr1_+_148603208
|
0.178
|
|
RPRD2
|
regulation of nuclear pre-mRNA domain containing 2
|
|
chr10_-_44794146
|
0.176
|
NM_007021
|
C10orf10
|
chromosome 10 open reading frame 10
|
|
chrX_+_70420194
|
0.175
|
|
NONO
|
non-POU domain containing, octamer-binding
|
|
chr1_-_147665783
|
0.175
|
|
HIST2H2BF
|
histone cluster 2, H2bf
|
|
chr19_+_44573858
|
0.171
|
|
MED29
|
mediator complex subunit 29
|
|
chr4_-_84254873
|
0.170
|
|
PLAC8
|
placenta-specific 8
|
|
chr6_-_30792830
|
0.170
|
|
MDC1
|
mediator of DNA-damage checkpoint 1
|
|
chr16_-_68880844
|
0.169
|
|
AARS
|
alanyl-tRNA synthetase
|
|
chr6_+_26212082
|
0.169
|
NM_003542
|
HIST1H4C
|
histone cluster 1, H4c
|
|
chr12_+_63090008
|
0.165
|
|
XPOT
|
exportin, tRNA (nuclear export receptor for tRNAs)
|
|
chr20_-_54400628
|
0.162
|
NM_003600
NM_198433
NM_198434
NM_198435
NM_198436
NM_198437
|
AURKA
|
aurora kinase A
|
|
chr1_-_180627023
|
0.160
|
NM_001033056
|
GLUL
|
glutamate-ammonia ligase
|
|
chr16_+_29728932
|
0.159
|
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor)
|
|
chr19_+_44573790
|
0.159
|
NM_017592
|
MED29
|
mediator complex subunit 29
|
|
chr16_-_68880908
|
0.158
|
NM_001605
|
AARS
|
alanyl-tRNA synthetase
|
|
chr8_+_107807415
|
0.158
|
NM_001198534
NM_001198535
|
OXR1
|
oxidation resistance 1
|
|
chr17_+_50697355
|
0.157
|
|
HLF
|
hepatic leukemia factor
|
|
chr18_-_58005159
|
0.156
|
|
PIGN
|
phosphatidylinositol glycan anchor biosynthesis, class N
|
|
chr3_+_173240126
|
0.156
|
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr16_+_80626331
|
0.156
|
NM_002153
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2
|
|
chr1_-_171442937
|
0.156
|
NM_003326
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4
|
|
chr6_+_111302679
|
0.155
|
NM_001634
NM_001033059
|
AMD1
|
adenosylmethionine decarboxylase 1
|
|
chr12_+_49728350
|
0.153
|
NM_001024668
NM_015416
|
LETMD1
|
LETM1 domain containing 1
|
|
chrX_+_70420221
|
0.151
|
|
NONO
|
non-POU domain containing, octamer-binding
|
|
chr1_-_58920315
|
0.151
|
|
|
|
|
chr16_-_57221240
|
0.150
|
NM_016284
NM_206999
|
CNOT1
|
CCR4-NOT transcription complex, subunit 1
|
|
chr5_-_149515331
|
0.150
|
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr16_-_57221235
|
0.149
|
|
CNOT1
|
CCR4-NOT transcription complex, subunit 1
|
|
chr6_-_26232109
|
0.149
|
NM_003526
|
HIST1H2BG
HIST1H2BC
|
histone cluster 1, H2bg
histone cluster 1, H2bc
|
|
chr14_-_57834558
|
0.148
|
|
FLJ31306
|
hypothetical LOC379025
|
|
chr14_+_95071136
|
0.148
|
|
GLRX5
|
glutaredoxin 5
|
|
chr16_+_80626402
|
0.147
|
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2
|
|
chr16_+_18986721
|
0.147
|
NM_001190983
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chr17_+_78009895
|
0.145
|
|
NARF
|
nuclear prelamin A recognition factor
|
|
chr2_-_218843088
|
0.144
|
|
AAMP
|
angio-associated, migratory cell protein
|
|
chrX_+_70420237
|
0.144
|
|
NONO
|
non-POU domain containing, octamer-binding
|
|
chr3_+_122109716
|
0.144
|
NM_014980
|
STXBP5L
|
syntaxin binding protein 5-like
|
|
chr17_-_44058612
|
0.142
|
NM_024017
|
HOXB9
|
homeobox B9
|
|
chr1_+_148603613
|
0.141
|
NM_015203
|
RPRD2
|
regulation of nuclear pre-mRNA domain containing 2
|
|
chr10_-_101180191
|
0.141
|
NM_002079
|
GOT1
|
glutamic-oxaloacetic transaminase 1, soluble (aspartate aminotransferase 1)
|
|
chr17_+_43480724
|
0.140
|
|
NFE2L1
|
nuclear factor (erythroid-derived 2)-like 1
|
|
chr19_+_46462125
|
0.140
|
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1
|
|
chr7_+_106897737
|
0.139
|
NM_005295
|
GPR22
|
G protein-coupled receptor 22
|
|
chr6_+_151603346
|
0.139
|
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr17_-_36074812
|
0.139
|
NM_152349
|
KRT222
|
keratin 222
|
|
chr18_-_50776276
|
0.138
|
NM_001143829
|
CCDC68
|
coiled-coil domain containing 68
|
|
chr5_+_145807059
|
0.137
|
NM_001040006
NM_006706
|
TCERG1
|
transcription elongation regulator 1
|
|
chr1_-_19101603
|
0.136
|
NM_003748
NM_170726
|
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1
|
|
chr14_-_73620826
|
0.135
|
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1
|
|
chr1_-_180621272
|
0.133
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr2_+_162908828
|
0.133
|
NM_012198
|
GCA
|
grancalcin, EF-hand calcium binding protein
|
|
chr1_-_57204208
|
0.132
|
NM_000066
|
C8B
|
complement component 8, beta polypeptide
|
|
chr7_-_143230096
|
0.130
|
NM_014719
|
FAM115A
|
family with sequence similarity 115, member A
|
|
chr17_+_43328514
|
0.130
|
NM_003110
|
SP2
|
Sp2 transcription factor
|
|
chr14_+_22860547
|
0.127
|
|
PABPN1
|
poly(A) binding protein, nuclear 1
|
|
chr4_+_6322506
|
0.127
|
|
WFS1
|
Wolfram syndrome 1 (wolframin)
|
|
chr2_+_24660849
|
0.126
|
NM_003743
NM_147223
NM_147233
|
NCOA1
|
nuclear receptor coactivator 1
|
|
chr14_-_73620912
|
0.126
|
NM_005589
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1
|
|
chrX_+_70420259
|
0.125
|
|
NONO
|
non-POU domain containing, octamer-binding
|
|
chr12_+_100512877
|
0.124
|
NM_002465
NM_206819
NM_206820
NM_206821
|
MYBPC1
|
myosin binding protein C, slow type
|
|
chr12_-_120715937
|
0.123
|
NM_019034
|
RHOF
|
ras homolog gene family, member F (in filopodia)
|
|
chr15_+_41264607
|
0.122
|
NM_012142
|
CCNDBP1
|
cyclin D-type binding-protein 1
|
|
chr4_+_155703581
|
0.121
|
NM_001184741
NM_005141
|
FGB
|
fibrinogen beta chain
|
|
chr20_+_29790564
|
0.121
|
NM_012112
|
TPX2
|
TPX2, microtubule-associated, homolog (Xenopus laevis)
|
|
chr14_+_22860511
|
0.120
|
|
PABPN1
|
poly(A) binding protein, nuclear 1
|
|
chr12_+_119609305
|
0.119
|
NM_014730
|
MLEC
|
malectin
|
|
chr9_-_83494198
|
0.118
|
|
TLE1
|
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila)
|
|
chr19_+_55398680
|
0.118
|
NM_001077186
NM_001145809
NM_024729
|
MYH14
|
myosin, heavy chain 14, non-muscle
|
|
chr10_-_115923874
|
0.117
|
|
C10orf118
|
chromosome 10 open reading frame 118
|
|
chr1_+_212843117
|
0.117
|
NM_016343
|
CENPF
|
centromere protein F, 350/400kDa (mitosin)
|
|
chr19_-_3928840
|
0.114
|
|
EEF2
|
eukaryotic translation elongation factor 2
|
|
chr5_-_89741233
|
0.113
|
NM_004365
|
CETN3
|
centrin, EF-hand protein, 3
|
|
chr5_-_147191428
|
0.113
|
NM_003122
|
SPINK1
|
serine peptidase inhibitor, Kazal type 1
|
|
chr3_-_123036526
|
0.111
|
|
IQCB1
|
IQ motif containing B1
|
|
chr12_-_6586811
|
0.110
|
NM_001273
|
CHD4
|
chromodomain helicase DNA binding protein 4
|
|
chr11_+_57121602
|
0.108
|
NM_000062
|
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1
|
|
chr14_+_57834962
|
0.108
|
NM_002892
NM_023000
NM_023001
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like)
|
|
chr5_+_43639044
|
0.107
|
|
NNT
|
nicotinamide nucleotide transhydrogenase
|
|
chr6_-_42821790
|
0.106
|
|
TBCC
|
tubulin folding cofactor C
|
|
chr12_+_6473519
|
0.106
|
|
NCAPD2
|
non-SMC condensin I complex, subunit D2
|
|
chr19_+_14501403
|
0.105
|
|
TECR
|
trans-2,3-enoyl-CoA reductase
|
|
chr1_-_148124738
|
0.104
|
NM_003528
|
HIST2H2BE
|
histone cluster 2, H2be
|
|
chr5_+_43638973
|
0.103
|
NM_182977
|
NNT
|
nicotinamide nucleotide transhydrogenase
|
|
chr10_+_114699953
|
0.103
|
NM_001146274
NM_001146283
NM_001146284
NM_001146285
NM_001146286
NM_001198525
NM_001198526
NM_001198527
NM_001198528
NM_001198529
NM_001198530
NM_001198531
NM_030756
|
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box)
|
|
chr10_+_76256305
|
0.103
|
NM_012330
|
MYST4
|
MYST histone acetyltransferase (monocytic leukemia) 4
|
|
chr3_+_52786642
|
0.102
|
NM_002215
|
ITIH1
|
inter-alpha (globulin) inhibitor H1
|
|
chr19_-_44032449
|
0.102
|
NM_001533
|
HNRNPL
|
heterogeneous nuclear ribonucleoprotein L
|
|
chr19_+_47416271
|
0.101
|
NM_133444
|
ZNF526
|
zinc finger protein 526
|
|
chr19_-_14108400
|
0.099
|
NM_018154
|
ASF1B
|
ASF1 anti-silencing function 1 homolog B (S. cerevisiae)
|
|
chr16_-_1816793
|
0.099
|
|
HAGH
|
hydroxyacylglutathione hydrolase
|
|
chr17_-_77411707
|
0.099
|
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide
|
|
chr19_-_14108267
|
0.098
|
|
ASF1B
|
ASF1 anti-silencing function 1 homolog B (S. cerevisiae)
|
|
chr17_+_16225117
|
0.097
|
|
UBB
|
ubiquitin B
|
|
chr17_-_77411659
|
0.097
|
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide
|
|
chr12_-_26877189
|
0.096
|
NM_002223
|
ITPR2
|
inositol 1,4,5-triphosphate receptor, type 2
|
|
chr4_+_129950222
|
0.096
|
NM_024900
NM_199320
|
PHF17
|
PHD finger protein 17
|
|
chr2_+_162908869
|
0.094
|
|
GCA
|
grancalcin, EF-hand calcium binding protein
|
|
chr19_+_14501406
|
0.094
|
|
TECR
|
trans-2,3-enoyl-CoA reductase
|
|
chr11_+_67984758
|
0.093
|
NM_001164160
NM_001164161
NM_001164162
NM_001164163
NM_001164164
NM_018312
|
PPP6R3
|
protein phosphatase 6, regulatory subunit 3
|
|
chr18_+_31074985
|
0.093
|
NM_001135178
NM_032347
|
ZNF397
|
zinc finger protein 397
|
|
chr2_+_27109341
|
0.093
|
|
TMEM214
|
transmembrane protein 214
|
|
chr5_+_139999182
|
0.093
|
NM_018502
|
TMCO6
|
transmembrane and coiled-coil domains 6
|
|
chr11_+_66140628
|
0.092
|
NM_001198845
NM_001198846
NM_001198836
NM_001198837
NM_006328
|
RBM14-RBM4
RBM14
|
RBM14-RBM4 readthrough
RNA binding motif protein 14
|
|
chr14_+_75197409
|
0.092
|
|
TTLL5
|
tubulin tyrosine ligase-like family, member 5
|
|
chr7_-_121571472
|
0.091
|
NM_005763
|
AASS
|
aminoadipate-semialdehyde synthase
|
|
chr5_+_43639069
|
0.090
|
|
NNT
|
nicotinamide nucleotide transhydrogenase
|
|
chr6_+_135544170
|
0.090
|
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr2_-_183439515
|
0.089
|
NM_001463
|
FRZB
|
frizzled-related protein
|
|
chr2_+_27109249
|
0.089
|
NM_001083590
NM_017727
|
TMEM214
|
transmembrane protein 214
|
|
chr17_-_77411803
|
0.089
|
NM_000918
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide
|
|
chr17_-_1534880
|
0.088
|
NM_006445
|
PRPF8
|
PRP8 pre-mRNA processing factor 8 homolog (S. cerevisiae)
|
|
chr7_-_135083906
|
0.088
|
NM_205855
|
FAM180A
|
family with sequence similarity 180, member A
|
|
chr17_-_1534848
|
0.087
|
|
PRPF8
|
PRP8 pre-mRNA processing factor 8 homolog (S. cerevisiae)
|
|
chr5_-_147191334
|
0.087
|
|
SPINK1
|
serine peptidase inhibitor, Kazal type 1
|
|
chr1_-_3702908
|
0.086
|
NM_020710
|
LRRC47
|
leucine rich repeat containing 47
|
|
chr4_-_140224794
|
0.086
|
|
ELF2
|
E74-like factor 2 (ets domain transcription factor)
|
|
chr12_-_118799457
|
0.086
|
NM_007174
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21)
|