|
chr8_-_13035102
|
2.259
|
NM_006094
|
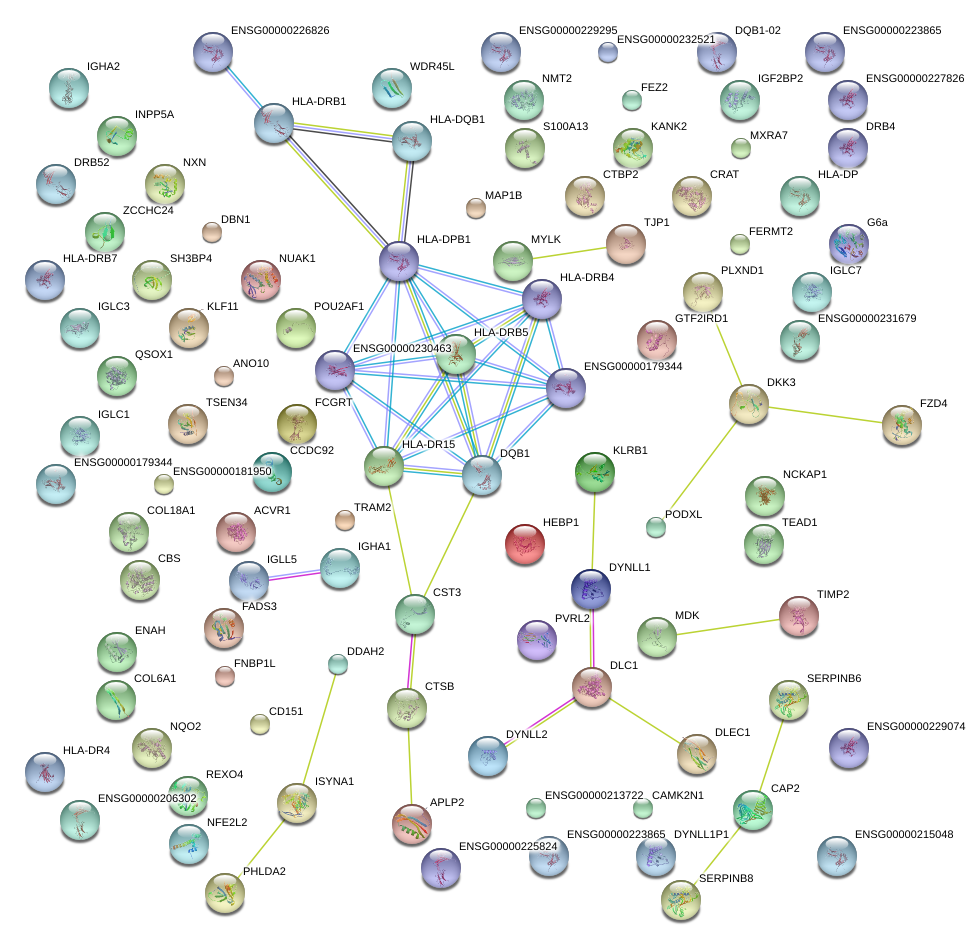
DLC1
|
deleted in liver cancer 1
|
|
chr3_-_187025501
|
1.734
|
NM_001007225
NM_006548
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chr11_+_46359878
|
1.601
|
NM_001012333
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr17_-_74432800
|
1.457
|
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr12_-_13044467
|
1.431
|
NM_015987
|
HEBP1
|
heme binding protein 1
|
|
chr11_+_46359855
|
1.380
|
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr11_+_46359781
|
1.367
|
NM_002391
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr11_-_2907169
|
1.362
|
NM_003311
|
PHLDA2
|
pleckstrin homology-like domain, family A, member 2
|
|
chr17_-_74432671
|
1.313
|
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr6_-_2916980
|
1.297
|
|
SERPINB6
|
serpin peptidase inhibitor, clade B (ovalbumin), member 6
|
|
chr17_-_72218661
|
1.283
|
|
MXRA7
|
matrix-remodelling associated 7
|
|
chr12_-_13044314
|
1.229
|
|
HEBP1
|
heme binding protein 1
|
|
chr6_-_31805547
|
1.221
|
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2
|
|
chr19_+_50041400
|
1.203
|
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B)
|
|
chr1_-_20685314
|
1.182
|
NM_018584
|
CAMK2N1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr1_-_223907283
|
1.178
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr12_-_105056577
|
1.172
|
|
NUAK1
|
NUAK family, SNF1-like kinase, 1
|
|
chr11_+_46359821
|
1.171
|
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr17_-_74433048
|
1.160
|
NM_003255
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr11_+_46359836
|
1.151
|
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr11_-_86343859
|
1.136
|
NM_012193
|
FZD4
|
frizzled homolog 4 (Drosophila)
|
|
chr2_+_10101081
|
1.126
|
NM_003597
NM_001177716
|
KLF11
|
Kruppel-like factor 11
|
|
chr12_-_123023052
|
1.110
|
NM_025140
|
CCDC92
|
coiled-coil domain containing 92
|
|
chr2_+_235525355
|
1.095
|
NM_014521
|
SH3BP4
|
SH3-domain binding protein 4
|
|
chr10_+_134201262
|
1.087
|
NM_005539
|
INPP5A
|
inositol polyphosphate-5-phosphatase, 40kDa
|
|
chr19_+_50041232
|
1.080
|
NM_001042724
NM_002856
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B)
|
|
chr10_-_15250616
|
1.076
|
NM_004808
|
NMT2
|
N-myristoyltransferase 2
|
|
chr10_-_80875097
|
1.063
|
NM_153367
|
ZCCHC24
|
zinc finger, CCHC domain containing 24
|
|
chr3_-_187025438
|
1.053
|
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chr19_+_50041424
|
1.024
|
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B)
|
|
chr14_-_52487558
|
1.024
|
NM_001134999
NM_001135000
NM_006832
|
FERMT2
|
fermitin family member 2
|
|
chr19_-_11169184
|
1.020
|
NM_001136191
|
KANK2
|
KN motif and ankyrin repeat domains 2
|
|
chr19_+_50041387
|
1.015
|
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B)
|
|
chr11_+_822965
|
0.986
|
|
CD151
|
CD151 molecule (Raph blood group)
|
|
chr6_+_2945025
|
0.984
|
NM_000904
|
NQO2
|
NAD(P)H dehydrogenase, quinone 2
|
|
chr11_+_823007
|
0.948
|
|
CD151
|
CD151 molecule (Raph blood group)
|
|
chr11_-_11987198
|
0.944
|
NM_015881
|
DKK3
|
dickkopf homolog 3 (Xenopus laevis)
|
|
chr21_+_45649429
|
0.942
|
NM_130445
|
COL18A1
|
collagen, type XVIII, alpha 1
|
|
chr5_+_71438796
|
0.927
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr11_-_11987115
|
0.911
|
|
DKK3
|
dickkopf homolog 3 (Xenopus laevis)
|
|
chr3_-_43638471
|
0.906
|
NM_018075
|
ANO10
|
anoctamin 10
|
|
chr22_-_34754343
|
0.903
|
NM_001082578
NM_001082579
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr3_-_43638425
|
0.902
|
|
ANO10
|
anoctamin 10
|
|
chr7_+_73506333
|
0.900
|
NM_005685
|
GTF2IRD1
|
GTF2I repeat domain containing 1
|
|
chr7_+_73506172
|
0.896
|
|
GTF2IRD1
|
GTF2I repeat domain containing 1
|
|
chr20_-_23566325
|
0.896
|
NM_000099
|
CST3
|
cystatin C
|
|
chr5_-_176832476
|
0.887
|
NM_080881
|
DBN1
|
drebrin 1
|
|
chr5_-_176833259
|
0.878
|
NM_004395
|
DBN1
|
drebrin 1
|
|
chr11_+_129445057
|
0.848
|
|
APLP2
|
amyloid beta (A4) precursor-like protein 2
|
|
chr11_-_11987414
|
0.848
|
|
DKK3
|
dickkopf homolog 3 (Xenopus laevis)
|
|
chr1_+_178390646
|
0.842
|
|
QSOX1
|
quiescin Q6 sulfhydryl oxidase 1
|
|
chr8_-_11762918
|
0.835
|
|
CTSB
|
cathepsin B
|
|
chr6_+_17501589
|
0.831
|
NM_006366
|
CAP2
|
CAP, adenylate cyclase-associated protein, 2 (yeast)
|
|
chr17_-_72218535
|
0.827
|
NM_001008528
NM_001008529
NM_198530
|
MXRA7
|
matrix-remodelling associated 7
|
|
chr22_+_21559959
|
0.815
|
NM_001178126
|
IGLL5
IGLC1
|
immunoglobulin lambda-like polypeptide 5
immunoglobulin lambda constant 1 (Mcg marker)
|
|
chr8_-_11763023
|
0.803
|
|
CTSB
|
cathepsin B
|
|
chr6_-_31805932
|
0.803
|
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2
|
|
chr3_-_125085834
|
0.802
|
NM_053025
NM_053026
NM_053027
NM_053028
|
MYLK
|
myosin light chain kinase
|
|
chr12_-_123023312
|
0.800
|
|
CCDC92
|
coiled-coil domain containing 92
|
|
chr3_-_130807865
|
0.795
|
NM_015103
|
PLXND1
|
plexin D1
|
|
chr21_+_45649719
|
0.787
|
|
|
|
|
chr5_-_176833197
|
0.780
|
|
DBN1
|
drebrin 1
|
|
chr6_-_31806012
|
0.780
|
NM_013974
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2
|
|
chr14_-_52487386
|
0.769
|
|
FERMT2
|
fermitin family member 2
|
|
chr14_-_106249999
|
0.764
|
|
IGHA1
|
immunoglobulin heavy constant alpha 1
|
|
chr11_+_46358909
|
0.764
|
NM_001012334
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr17_-_78199384
|
0.752
|
NM_019613
|
WDR45L
|
WDR45-like
|
|
chr1_+_178390636
|
0.738
|
|
QSOX1
|
quiescin Q6 sulfhydryl oxidase 1
|
|
chr11_+_12652427
|
0.736
|
NM_021961
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor)
|
|
chr2_-_36678657
|
0.731
|
NM_001042548
NM_005102
|
FEZ2
|
fasciculation and elongation protein zeta 2 (zygin II)
|
|
chr11_+_822909
|
0.729
|
NM_001039490
NM_004357
NM_139029
NM_139030
|
CD151
|
CD151 molecule (Raph blood group)
|
|
chr22_+_21340757
|
0.726
|
|
|
|
|
chr9_-_130912728
|
0.719
|
NM_000755
|
CRAT
|
carnitine O-acetyltransferase
|
|
chr1_+_178390529
|
0.714
|
NM_001004128
NM_002826
|
QSOX1
|
quiescin Q6 sulfhydryl oxidase 1
|
|
chr21_-_43368963
|
0.714
|
NM_000071
|
CBS
|
cystathionine-beta-synthase
|
|
chr11_-_61415429
|
0.709
|
NM_021727
|
FADS3
|
fatty acid desaturase 3
|
|
chr17_-_829655
|
0.706
|
NM_022463
|
NXN
|
nucleoredoxin
|
|
chr1_-_151873191
|
0.704
|
NM_001024210
|
S100A13
|
S100 calcium binding protein A13
|
|
chr1_+_178390654
|
0.698
|
|
QSOX1
|
quiescin Q6 sulfhydryl oxidase 1
|
|
chr19_+_54708247
|
0.697
|
NM_004107
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chr6_-_32665409
|
0.697
|
NM_002124
|
HLA-DRB1
HLA-DRB4
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 1
major histocompatibility complex, class II, DR beta 4
major histocompatibility complex, class II, DR beta 5
|
|
chr8_-_11763004
|
0.692
|
|
CTSB
|
cathepsin B
|
|
chr1_+_178390660
|
0.687
|
|
QSOX1
|
quiescin Q6 sulfhydryl oxidase 1
|
|
chr7_+_73506047
|
0.687
|
NM_016328
|
GTF2IRD1
|
GTF2I repeat domain containing 1
|
|
chr11_+_129444877
|
0.685
|
NM_001142276
NM_001142277
NM_001142278
NM_001642
|
APLP2
|
amyloid beta (A4) precursor-like protein 2
|
|
chr19_+_54708312
|
0.683
|
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chr21_+_46226072
|
0.683
|
NM_001848
|
COL6A1
|
collagen, type VI, alpha 1
|
|
chr2_-_183610992
|
0.678
|
|
NCKAP1
|
NCK-associated protein 1
|
|
chr2_-_158440587
|
0.675
|
NM_001111067
|
ACVR1
|
activin A receptor, type I
|
|
chr6_-_52549673
|
0.671
|
|
TRAM2
|
translocation associated membrane protein 2
|
|
chr10_-_126839000
|
0.666
|
|
CTBP2
|
C-terminal binding protein 2
|
|
chr15_-_27901828
|
0.666
|
NM_003257
NM_175610
|
TJP1
|
tight junction protein 1 (zona occludens 1)
|
|
chr22_+_21116283
|
0.665
|
|
|
|
|
chr19_+_59385930
|
0.664
|
NM_024075
|
TSEN34
|
tRNA splicing endonuclease 34 homolog (S. cerevisiae)
|
|
chr1_+_93686154
|
0.662
|
NM_001024948
NM_001164473
NM_017737
|
FNBP1L
|
formin binding protein 1-like
|
|
chr7_-_130891861
|
0.658
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr6_+_33151703
|
0.653
|
NM_002121
|
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1
|
|
chr3_-_45242808
|
0.653
|
NM_015444
|
TMEM158
|
transmembrane protein 158 (gene/pseudogene)
|
|
chr11_-_110755112
|
0.653
|
|
POU2AF1
|
POU class 2 associating factor 1
|
|
chr9_-_130912603
|
0.652
|
|
CRAT
|
carnitine O-acetyltransferase
|
|
chr6_-_2916786
|
0.652
|
|
SERPINB6
|
serpin peptidase inhibitor, clade B (ovalbumin), member 6
|
|
chr19_-_18409788
|
0.651
|
NM_001170939
|
ISYNA1
|
inositol-3-phosphate synthase 1
|
|
chr8_-_11763031
|
0.650
|
|
CTSB
|
cathepsin B
|
|
chr10_-_126839056
|
0.646
|
NM_001329
|
CTBP2
|
C-terminal binding protein 2
|
|
chr19_+_3045528
|
0.636
|
|
GNA11
|
guanine nucleotide binding protein (G protein), alpha 11 (Gq class)
|
|
chr3_-_45242624
|
0.635
|
|
TMEM158
|
transmembrane protein 158 (gene/pseudogene)
|
|
chr11_+_129444997
|
0.634
|
|
APLP2
|
amyloid beta (A4) precursor-like protein 2
|
|
chr1_-_22136263
|
0.629
|
NM_005529
|
HSPG2
|
heparan sulfate proteoglycan 2
|
|
chr5_+_141468498
|
0.627
|
NM_030571
|
NDFIP1
|
Nedd4 family interacting protein 1
|
|
chr4_-_158111995
|
0.627
|
NM_016205
|
PDGFC
|
platelet derived growth factor C
|
|
chr2_-_218551786
|
0.624
|
|
TNS1
|
tensin 1
|
|
chrX_-_101658297
|
0.621
|
NM_021992
|
TMSB15A
TMSB15B
|
thymosin beta 15a
thymosin beta 15B
|
|
chr11_-_2863536
|
0.620
|
NM_000076
NM_001122630
NM_001122631
|
CDKN1C
|
cyclin-dependent kinase inhibitor 1C (p57, Kip2)
|
|
chr15_-_20637731
|
0.618
|
NM_144599
|
NIPA1
|
non imprinted in Prader-Willi/Angelman syndrome 1
|
|
chr1_-_225572419
|
0.618
|
NM_003607
NM_014826
|
CDC42BPA
|
CDC42 binding protein kinase alpha (DMPK-like)
|
|
chr14_-_105163301
|
0.616
|
|
IGHG4
|
immunoglobulin heavy constant gamma 4 (G4m marker)
|
|
chr1_+_178390598
|
0.615
|
|
QSOX1
|
quiescin Q6 sulfhydryl oxidase 1
|
|
chr7_-_143230096
|
0.615
|
NM_014719
|
FAM115A
|
family with sequence similarity 115, member A
|
|
chr22_+_21370394
|
0.611
|
|
IGL@
|
immunoglobulin lambda locus
|
|
chr6_-_33149244
|
0.611
|
NM_033554
|
HLA-DPA1
|
major histocompatibility complex, class II, DP alpha 1
|
|
chr1_-_52228880
|
0.609
|
NM_002867
|
RAB3B
|
RAB3B, member RAS oncogene family
|
|
chr3_+_156280338
|
0.605
|
NM_007287
|
MME
|
membrane metallo-endopeptidase
|
|
chr4_-_177950658
|
0.603
|
NM_005429
|
VEGFC
|
vascular endothelial growth factor C
|
|
chr11_+_129445017
|
0.599
|
|
APLP2
|
amyloid beta (A4) precursor-like protein 2
|
|
chrX_-_34585287
|
0.597
|
NM_031442
|
TMEM47
|
transmembrane protein 47
|
|
chr3_+_123526700
|
0.596
|
NM_005213
|
CSTA
|
cystatin A (stefin A)
|
|
chr14_-_105182025
|
0.587
|
|
|
|
|
chr12_+_6424293
|
0.586
|
NM_001242
|
CD27
|
CD27 molecule
|
|
chr19_-_54835162
|
0.585
|
NM_006270
|
RRAS
|
related RAS viral (r-ras) oncogene homolog
|
|
chr10_+_94439658
|
0.585
|
NM_002729
|
HHEX
|
hematopoietically expressed homeobox
|
|
chr16_-_63713485
|
0.582
|
|
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast)
|
|
chr10_-_725522
|
0.582
|
NM_014974
|
DIP2C
|
DIP2 disco-interacting protein 2 homolog C (Drosophila)
|
|
chr2_+_24126140
|
0.579
|
|
FKBP1B
|
FK506 binding protein 1B, 12.6 kDa
|
|
chr1_+_181259317
|
0.579
|
|
|
|
|
chr18_+_3439972
|
0.577
|
NM_173209
NM_173208
NM_003244
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr14_-_105181939
|
0.575
|
|
IGHG2
|
immunoglobulin heavy constant gamma 2 (G2m marker)
|
|
chr14_-_68515791
|
0.570
|
NM_001102
NM_001130004
NM_001130005
|
ACTN1
|
actinin, alpha 1
|
|
chr5_-_179166378
|
0.570
|
NM_014275
|
MGAT4B
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B
|
|
chr17_-_1566223
|
0.570
|
|
C17orf91
|
chromosome 17 open reading frame 91
|
|
chr1_+_243200275
|
0.569
|
|
EFCAB2
|
EF-hand calcium binding domain 2
|
|
chr9_-_73573121
|
0.569
|
NM_001135820
NM_013390
|
TMEM2
|
transmembrane protein 2
|
|
chr14_-_105308639
|
0.567
|
|
|
|
|
chr11_-_11987458
|
0.566
|
NM_013253
|
DKK3
|
dickkopf homolog 3 (Xenopus laevis)
|
|
chr7_+_93861808
|
0.565
|
NM_000089
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr2_+_24126087
|
0.563
|
NM_004116
NM_054033
|
FKBP1B
|
FK506 binding protein 1B, 12.6 kDa
|
|
chr1_+_181258952
|
0.561
|
NM_002293
|
LAMC1
|
laminin, gamma 1 (formerly LAMB2)
|
|
chr18_-_24010944
|
0.561
|
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal)
|
|
chr1_-_95165037
|
0.555
|
|
CNN3
|
calponin 3, acidic
|
|
chr19_+_3045266
|
0.555
|
NM_002067
|
GNA11
|
guanine nucleotide binding protein (G protein), alpha 11 (Gq class)
|
|
chr16_-_87450734
|
0.553
|
|
GALNS
|
galactosamine (N-acetyl)-6-sulfate sulfatase
|
|
chr14_-_105308720
|
0.553
|
|
IGHM
|
immunoglobulin heavy constant mu
|
|
chr7_-_1465487
|
0.548
|
|
MICALL2
|
MICAL-like 2
|
|
chr2_+_20510312
|
0.547
|
NM_004040
|
RHOB
|
ras homolog gene family, member B
|
|
chr17_-_60088637
|
0.544
|
|
SMURF2
|
SMAD specific E3 ubiquitin protein ligase 2
|
|
chr21_-_43369492
|
0.543
|
NM_001178008
NM_001178009
|
CBS
|
cystathionine-beta-synthase
|
|
chr14_-_105280425
|
0.540
|
|
IGHG1
|
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr3_-_15875932
|
0.534
|
NM_015199
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr7_-_50828608
|
0.533
|
NM_001001555
|
GRB10
|
growth factor receptor-bound protein 10
|
|
chr14_-_68515525
|
0.532
|
|
ACTN1
|
actinin, alpha 1
|
|
chr3_+_12813170
|
0.532
|
NM_001162499
NM_012298
|
CAND2
|
cullin-associated and neddylation-dissociated 2 (putative)
|
|
chr4_-_1156533
|
0.532
|
NM_012445
|
SPON2
|
spondin 2, extracellular matrix protein
|
|
chr20_+_30813851
|
0.530
|
NM_006892
NM_175848
NM_175849
|
DNMT3B
|
DNA (cytosine-5-)-methyltransferase 3 beta
|
|
chr17_-_1335763
|
0.529
|
NM_001080950
|
MYO1C
|
myosin IC
|
|
chr10_-_62373828
|
0.529
|
NM_014836
|
RHOBTB1
|
Rho-related BTB domain containing 1
|
|
chr14_+_95575319
|
0.523
|
|
C14orf132
|
chromosome 14 open reading frame 132
|
|
chr6_-_52549774
|
0.522
|
NM_012288
|
TRAM2
|
translocation associated membrane protein 2
|
|
chr6_+_37245860
|
0.521
|
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr19_-_60350122
|
0.521
|
|
TNNT1
|
troponin T type 1 (skeletal, slow)
|
|
chr11_+_129445002
|
0.520
|
|
APLP2
|
amyloid beta (A4) precursor-like protein 2
|
|
chr19_+_54708302
|
0.519
|
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chr14_+_105012106
|
0.518
|
NM_001312
|
CRIP2
|
cysteine-rich protein 2
|
|
chr10_+_121400741
|
0.515
|
NM_004281
|
BAG3
|
BCL2-associated athanogene 3
|
|
chr14_-_105182159
|
0.513
|
|
IGHG2
|
immunoglobulin heavy constant gamma 2 (G2m marker)
|
|
chr20_+_11819364
|
0.512
|
NM_181443
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr11_-_65396775
|
0.511
|
NM_016938
|
EFEMP2
|
EGF containing fibulin-like extracellular matrix protein 2
|
|
chrX_+_54852217
|
0.511
|
NM_201222
|
MAGED2
|
melanoma antigen family D, 2
|
|
chr22_-_35875890
|
0.509
|
NM_000878
|
IL2RB
|
interleukin 2 receptor, beta
|
|
chr8_-_11763037
|
0.507
|
NM_001908
NM_147780
NM_147781
NM_147782
NM_147783
|
CTSB
|
cathepsin B
|
|
chr12_-_51132129
|
0.507
|
NM_005555
|
KRT6B
|
keratin 6B
|
|
chr17_+_77528699
|
0.506
|
NM_024083
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1
|
|
chr5_-_179431204
|
0.505
|
|
RNF130
|
ring finger protein 130
|
|
chr9_+_102275455
|
0.505
|
|
TMEFF1
|
transmembrane protein with EGF-like and two follistatin-like domains 1
|
|
chrX_-_153428621
|
0.502
|
|
G6PD
|
glucose-6-phosphate dehydrogenase
|
|
chr5_-_16989371
|
0.499
|
NM_012334
|
MYO10
|
myosin X
|
|
chrX_-_17789276
|
0.497
|
NM_001172732
NM_001172739
NM_001172743
NM_021785
|
RAI2
|
retinoic acid induced 2
|
|
chr14_-_64508547
|
0.497
|
NM_198686
|
RAB15
|
RAB15, member RAS onocogene family
|
|
chr5_+_141468569
|
0.495
|
|
NDFIP1
|
Nedd4 family interacting protein 1
|
|
chr16_-_63713382
|
0.494
|
NM_001797
|
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast)
|
|
chrX_+_9392980
|
0.493
|
NM_005647
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr22_+_21495273
|
0.493
|
|
|
|
|
chr7_-_150847529
|
0.493
|
|
RHEB
|
Ras homolog enriched in brain
|
|
chr17_-_38427853
|
0.493
|
|
VAT1
|
vesicle amine transport protein 1 homolog (T. californica)
|
|
chr8_-_25371858
|
0.492
|
NM_017634
|
KCTD9
|
potassium channel tetramerisation domain containing 9
|
|
chr19_-_60350118
|
0.491
|
|
TNNT1
|
troponin T type 1 (skeletal, slow)
|
|
chr1_-_84928473
|
0.488
|
NM_001166295
NM_001166417
NM_001166293
NM_001166294
NM_014021
|
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein
|
|
chr14_-_29466562
|
0.487
|
NM_002742
|
PRKD1
|
protein kinase D1
|
|
chr19_-_59385265
|
0.487
|
|
MBOAT7
|
membrane bound O-acyltransferase domain containing 7
|
|
chr16_-_63713466
|
0.486
|
|
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast)
|
|
chr2_+_46779553
|
0.486
|
NM_014011
|
SOCS5
|
suppressor of cytokine signaling 5
|
|
chr17_-_38427928
|
0.485
|
|
VAT1
|
vesicle amine transport protein 1 homolog (T. californica)
|