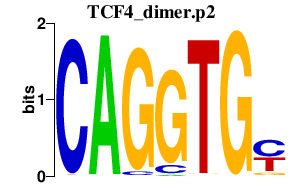
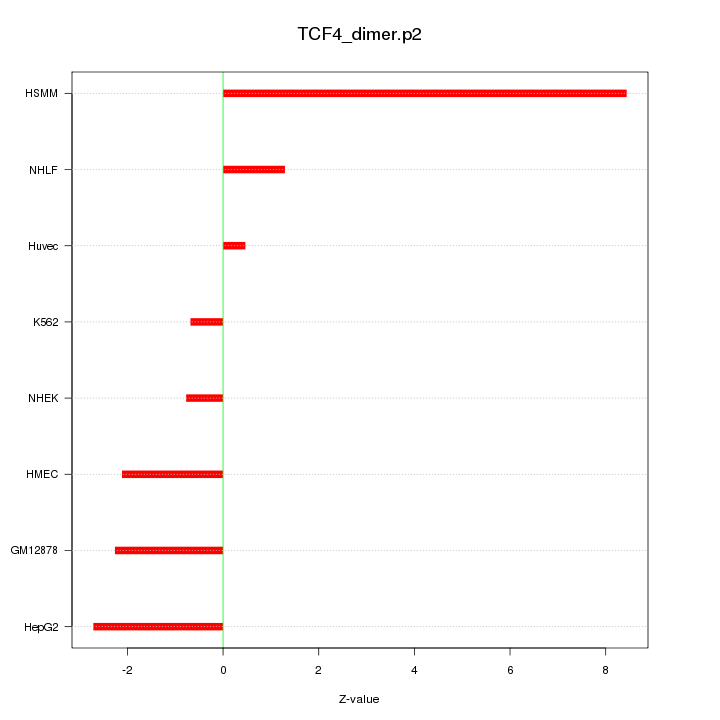
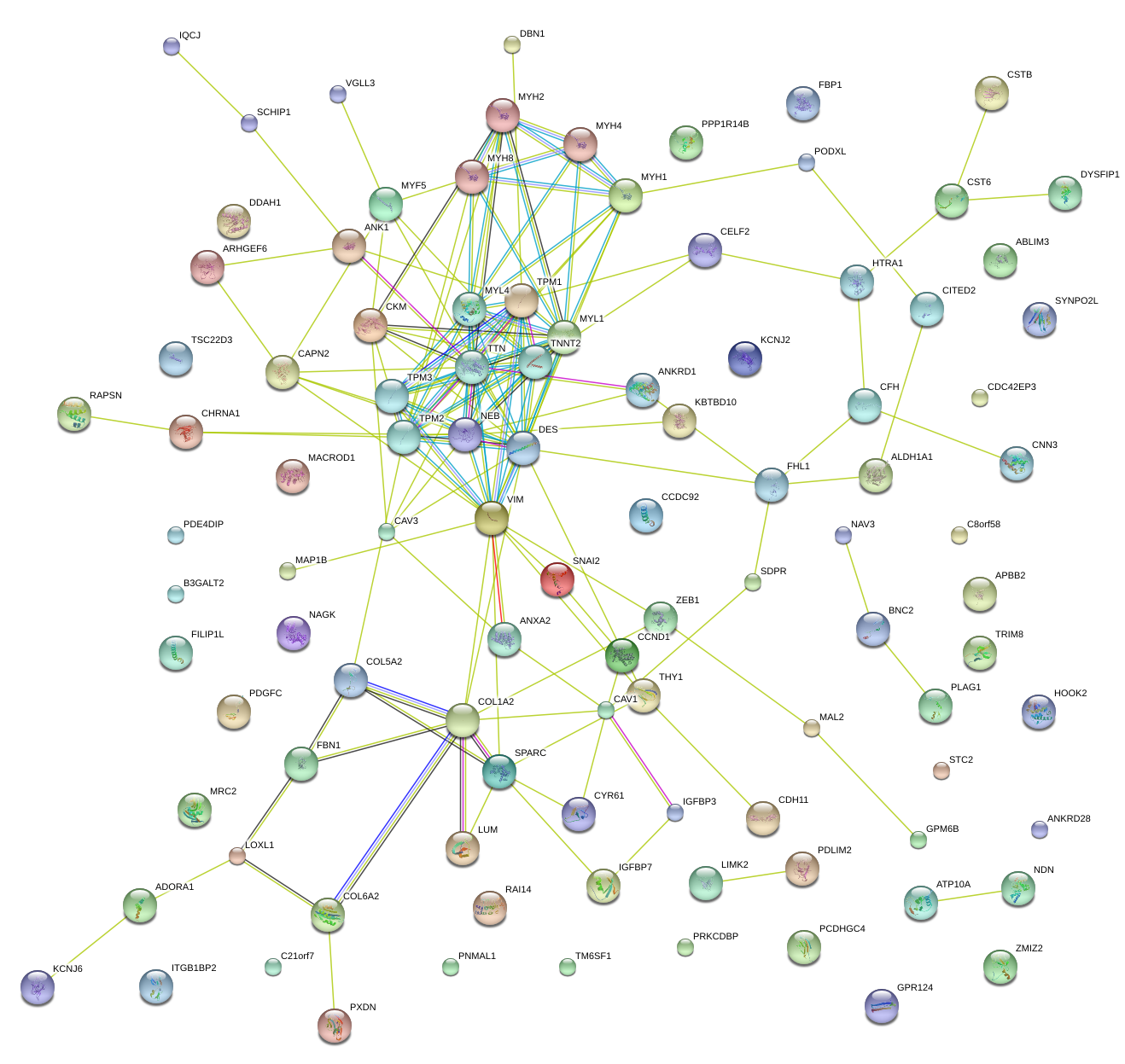
|
chr15_+_72006015
|
8.915
|
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr9_-_35679799
|
6.343
|
|
TPM2
|
tropomyosin 2 (beta)
|
|
chr9_-_35679902
|
6.126
|
NM_003289
NM_213674
|
TPM2
|
tropomyosin 2 (beta)
|
|
chr15_+_72005847
|
5.759
|
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr15_+_72005839
|
5.661
|
NM_005576
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr15_-_46725087
|
5.501
|
NM_000138
|
FBN1
|
fibrillin 1
|
|
chr4_-_81213207
|
5.401
|
|
|
|
|
chr2_+_219991342
|
4.868
|
NM_001927
|
DES
|
desmin
|
|
chr9_-_74757735
|
4.591
|
NM_000689
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1
|
|
chr8_-_49996353
|
3.964
|
NM_003068
|
SNAI2
|
snail homolog 2 (Drosophila)
|
|
chr7_+_115953582
|
3.937
|
NM_001172896
NM_001172897
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr7_-_45927263
|
3.868
|
NM_000598
NM_001013398
|
IGFBP3
|
insulin-like growth factor binding protein 3
|
|
chr2_+_170074457
|
3.831
|
NM_006063
|
KBTBD10
|
kelch repeat and BTB (POZ) domain containing 10
|
|
chr6_-_139737042
|
3.757
|
NM_001168389
NM_001168388
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2
|
|
chr3_-_87122936
|
3.536
|
NM_016206
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr21_+_46355797
|
3.490
|
|
COL6A2
|
collagen, type VI, alpha 2
|
|
chr7_-_130891861
|
3.406
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr11_+_65536037
|
3.278
|
NM_001323
|
CST6
|
cystatin E/M
|
|
chr2_-_175337386
|
3.079
|
NM_000079
NM_001039523
|
CHRNA1
|
cholinergic receptor, nicotinic, alpha 1 (muscle)
|
|
chr15_+_72005875
|
3.005
|
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr12_+_76749199
|
2.991
|
NM_014903
|
NAV3
|
neuron navigator 3
|
|
chr12_-_90029328
|
2.991
|
|
LUM
|
lumican
|
|
chr4_-_57671273
|
2.974
|
NM_001553
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chr15_+_72005864
|
2.974
|
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr2_-_192420168
|
2.946
|
NM_004657
|
SDPR
|
serum deprivation response
|
|
chr4_-_57671307
|
2.939
|
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chr12_-_90029672
|
2.862
|
NM_002345
|
LUM
|
lumican
|
|
chr10_+_31648103
|
2.859
|
NM_001174093
NM_001174095
NM_001174096
NM_030751
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr10_-_92671005
|
2.856
|
NM_014391
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr2_-_189752575
|
2.854
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr10_+_11100111
|
2.848
|
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr4_-_40911212
|
2.845
|
NM_001166050
NM_004307
NM_173075
|
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2
|
|
chr21_+_29439768
|
2.814
|
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chr2_-_192419896
|
2.812
|
|
SDPR
|
serum deprivation response
|
|
chr4_-_57671112
|
2.792
|
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chr5_+_148501178
|
2.750
|
NM_014945
|
ABLIM3
|
actin binding LIM protein family, member 3
|
|
chr1_+_221955876
|
2.735
|
NM_001146068
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr1_+_181259317
|
2.731
|
|
|
|
|
chrX_-_13866451
|
2.718
|
|
GPM6B
|
glycoprotein M6B
|
|
chrX_-_135691168
|
2.665
|
NM_004840
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr2_-_37752731
|
2.663
|
NM_006449
|
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3
|
|
chr8_+_22492586
|
2.650
|
|
PDLIM2
|
PDZ and LIM domain 2 (mystique)
|
|
chr11_-_47427263
|
2.616
|
NM_005055
NM_032645
|
RAPSN
|
receptor-associated protein of the synapse
|
|
chr10_+_104394185
|
2.590
|
NM_030912
|
TRIM8
|
tripartite motif containing 8
|
|
chr17_+_42641712
|
2.586
|
NM_002476
|
MYL4
|
myosin, light chain 4, alkali; atrial, embryonic
|
|
chr12_+_79634820
|
2.578
|
NM_005593
|
MYF5
|
myogenic factor 5
|
|
chr1_-_95165089
|
2.557
|
NM_001839
|
CNN3
|
calponin 3, acidic
|
|
chr10_-_92670764
|
2.547
|
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr7_+_93861808
|
2.519
|
NM_000089
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr10_+_11099860
|
2.491
|
NM_006561
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr10_+_31647425
|
2.490
|
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr11_-_6298284
|
2.480
|
NM_145040
|
PRKCDBP
|
protein kinase C, delta binding protein
|
|
chr15_+_61127367
|
2.444
|
NM_001018008
|
TPM1
|
tropomyosin 1 (alpha)
|
|
chr6_-_139737432
|
2.444
|
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2
|
|
chr6_-_139737477
|
2.440
|
NM_006079
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2
|
|
chr7_+_93862137
|
2.427
|
|
COL1A2
PLAG1
|
collagen, type I, alpha 2
pleiomorphic adenoma gene 1
|
|
chr4_-_158111995
|
2.396
|
NM_016205
|
PDGFC
|
platelet derived growth factor C
|
|
chr1_-_199613427
|
2.372
|
NM_000364
NM_001001430
NM_001001431
NM_001001432
|
TNNT2
|
troponin T type 2 (cardiac)
|
|
chr10_+_11100075
|
2.370
|
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr1_-_199613406
|
2.250
|
|
TNNT2
|
troponin T type 2 (cardiac)
|
|
chr2_-_210888010
|
2.200
|
NM_079420
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast
|
|
chrX_-_106905545
|
2.197
|
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr3_-_15875932
|
2.177
|
NM_015199
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr1_-_85703198
|
2.173
|
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr9_+_33614214
|
2.098
|
|
ANXA2P2
|
annexin A2 pseudogene 2
|
|
chr3_-_101316038
|
2.084
|
NM_001042459
NM_182909
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chrX_+_70438352
|
2.081
|
NM_012278
|
ITGB1BP2
|
integrin beta 1 binding protein (melusin) 2
|
|
chr3_+_160269734
|
1.986
|
NM_001197113
NM_001197114
NM_001042705
NM_001042706
NM_001197100
|
IQCJ-SCHIP1
IQCJ
|
IQCJ-SCHIP1 readthrough
IQ motif containing J
|
|
chr11_-_118799452
|
1.978
|
NM_006288
|
THY1
|
Thy-1 cell surface antigen
|
|
chr1_-_143643064
|
1.976
|
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr10_-_75080784
|
1.974
|
NM_024875
|
SYNPO2L
|
synaptopodin 2-like
|
|
chr19_-_12747263
|
1.950
|
|
HOOK2
|
hook homolog 2 (Drosophila)
|
|
chr1_-_191422346
|
1.948
|
NM_003783
|
B3GALT2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2
|
|
chr12_-_123023312
|
1.938
|
|
CCDC92
|
coiled-coil domain containing 92
|
|
chr16_-_63713382
|
1.937
|
NM_001797
|
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast)
|
|
chr10_+_124210980
|
1.924
|
NM_002775
|
HTRA1
|
HtrA serine peptidase 1
|
|
chr4_-_158111504
|
1.913
|
|
PDGFC
|
platelet derived growth factor C
|
|
chr9_-_96441409
|
1.879
|
|
FBP1
|
fructose-1,6-bisphosphatase 1
|
|
chr5_+_140844810
|
1.849
|
NM_018928
NM_032406
|
PCDHGC4
|
protocadherin gamma subfamily C, 4
|
|
chr17_+_58058774
|
1.839
|
|
MRC2
|
mannose receptor, C type 2
|
|
chr3_+_8750485
|
1.829
|
NM_033337
NM_001234
|
CAV3
|
caveolin 3
|
|
chr5_-_151046680
|
1.815
|
NM_003118
|
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin)
|
|
chr2_+_71149193
|
1.804
|
|
NAGK
|
N-acetylglucosamine kinase
|
|
chr5_-_176832476
|
1.789
|
NM_080881
|
DBN1
|
drebrin 1
|
|
chr16_-_63713466
|
1.788
|
|
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast)
|
|
chr8_+_120289735
|
1.787
|
NM_052886
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene)
|
|
chr16_-_63713485
|
1.781
|
|
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast)
|
|
chr19_-_51666629
|
1.767
|
NM_001103149
NM_018215
|
PNMAL1
|
PNMA-like 1
|
|
chr15_+_81567327
|
1.753
|
NM_001144903
NM_023003
|
TM6SF1
|
transmembrane 6 superfamily member 1
|
|
chr19_-_50517971
|
1.742
|
|
CKM
|
creatine kinase, muscle
|
|
chr8_-_41641881
|
1.736
|
NM_001142445
NM_020478
NM_020480
|
ANK1
|
ankyrin 1, erythrocytic
|
|
chr11_+_69165018
|
1.725
|
NM_053056
|
CCND1
|
cyclin D1
|
|
chr11_-_63770957
|
1.721
|
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B
|
|
chr1_+_85819004
|
1.720
|
NM_001554
|
CYR61
|
cysteine-rich, angiogenic inducer, 61
|
|
chr1_+_201364050
|
1.715
|
|
ADORA1
|
adenosine A1 receptor
|
|
chr10_+_11247059
|
1.712
|
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr8_+_37773991
|
1.712
|
|
GPR124
|
G protein-coupled receptor 124
|
|
chrX_-_106905657
|
1.712
|
NM_198057
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr15_-_23659329
|
1.707
|
NM_024490
|
ATP10A
|
ATPase, class V, type 10A
|
|
chrX_-_13866565
|
1.697
|
NM_001001994
|
GPM6B
|
glycoprotein M6B
|
|
chr17_+_65677255
|
1.690
|
NM_000891
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2
|
|
chr10_+_124211353
|
1.682
|
|
HTRA1
|
HtrA serine peptidase 1
|
|
chr2_-_179380301
|
1.666
|
NM_003319
NM_133378
NM_133379
NM_133432
NM_133437
|
TTN
|
titin
|
|
chr11_-_118799058
|
1.643
|
|
THY1
|
Thy-1 cell surface antigen
|
|
chr10_+_17311347
|
1.633
|
|
VIM
|
vimentin
|
|
chrX_+_135079120
|
1.614
|
NM_001159704
|
FHL1
|
four and a half LIM domains 1
|
|
chr11_-_63689780
|
1.609
|
|
MACROD1
|
MACRO domain containing 1
|
|
chr2_-_152299229
|
1.607
|
NM_001164507
NM_001164508
NM_004543
|
NEB
|
nebulin
|
|
chr17_-_10393664
|
1.602
|
NM_001100112
NM_017534
|
MYH2
|
myosin, heavy chain 2, skeletal muscle, adult
|
|
chr15_-_21483457
|
1.592
|
|
NDN
|
necdin homolog (mouse)
|
|
chr5_+_71438796
|
1.588
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr21_-_38210531
|
1.583
|
NM_002240
|
KCNJ6
|
potassium inwardly-rectifying channel, subfamily J, member 6
|
|
chr7_+_44754978
|
1.581
|
NM_031449
|
ZMIZ2
|
zinc finger, MIZ-type containing 2
|
|
chr19_-_51666503
|
1.571
|
|
PNMAL1
|
PNMA-like 1
|
|
chr10_+_11246934
|
1.568
|
NM_001025076
NM_001083591
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr2_-_1727324
|
1.562
|
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr11_-_63690069
|
1.561
|
NM_014067
|
MACROD1
|
MACRO domain containing 1
|
|
chr10_+_17311788
|
1.559
|
|
VIM
|
vimentin
|
|
chr9_-_16717887
|
1.549
|
|
BNC2
|
basonuclin 2
|
|
chr5_+_34692239
|
1.540
|
|
RAI14
|
retinoic acid induced 14
|
|
chr5_-_172687798
|
1.530
|
|
STC2
|
stanniocalcin 2
|
|
chr19_-_12747421
|
1.522
|
NM_001100176
NM_013312
|
HOOK2
|
hook homolog 2 (Drosophila)
|
|
chr11_-_2108035
|
1.515
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr10_+_17311962
|
1.513
|
|
VIM
|
vimentin
|
|
chr19_-_11169184
|
1.511
|
NM_001136191
|
KANK2
|
KN motif and ankyrin repeat domains 2
|
|
chr21_+_25933459
|
1.509
|
NM_021219
|
JAM2
|
junctional adhesion molecule 2
|
|
chr17_+_45488356
|
1.482
|
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor)
|
|
chr16_+_87765661
|
1.478
|
NM_004933
|
CDH15
|
cadherin 15, type 1, M-cadherin (myotubule)
|
|
chr2_-_1727247
|
1.475
|
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr2_+_223625105
|
1.470
|
NM_080671
|
KCNE4
|
potassium voltage-gated channel, Isk-related family, member 4
|
|
chr3_-_180272229
|
1.470
|
|
ZMAT3
|
zinc finger, matrin-type 3
|
|
chr11_+_86189138
|
1.460
|
NM_007173
|
PRSS23
|
protease, serine, 23
|
|
chrX_-_13866366
|
1.457
|
|
|
|
|
chrX_+_48221323
|
1.451
|
NM_177434
|
FTSJ1
|
FtsJ homolog 1 (E. coli)
|
|
chr17_+_45488330
|
1.449
|
NM_002204
NM_005501
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor)
|
|
chr8_-_122722674
|
1.448
|
NM_005328
|
HAS2
|
hyaluronan synthase 2
|
|
chrX_-_135690770
|
1.440
|
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr11_-_63770840
|
1.422
|
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B
|
|
chrX_-_135690699
|
1.413
|
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr1_+_144236346
|
1.403
|
NM_003637
|
ITGA10
|
integrin, alpha 10
|
|
chr11_+_12088639
|
1.397
|
NM_014632
|
MICAL2
|
microtubule associated monoxygenase, calponin and LIM domain containing 2
|
|
chr2_-_1727292
|
1.391
|
NM_012293
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr1_-_32940943
|
1.384
|
NM_001161708
NM_030786
|
SYNC
|
syncoilin, intermediate filament protein
|
|
chr4_+_40953698
|
1.378
|
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr6_-_46566969
|
1.376
|
|
RCAN2
|
regulator of calcineurin 2
|
|
chr20_+_19818209
|
1.368
|
NM_018993
|
RIN2
|
Ras and Rab interactor 2
|
|
chr6_+_129245978
|
1.365
|
NM_000426
NM_001079823
|
LAMA2
|
laminin, alpha 2
|
|
chr1_-_109386112
|
1.357
|
NM_001142550
NM_001142551
NM_014969
|
WDR47
|
WD repeat domain 47
|
|
chr15_-_53996597
|
1.347
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr5_+_34692352
|
1.339
|
NM_001145520
|
RAI14
|
retinoic acid induced 14
|
|
chr3_-_180272291
|
1.324
|
NM_022470
NM_152240
|
ZMAT3
|
zinc finger, matrin-type 3
|
|
chr11_+_1816808
|
1.311
|
NM_003282
|
TNNI2
|
troponin I type 2 (skeletal, fast)
|
|
chr17_-_15109315
|
1.309
|
NM_000304
|
PMP22
|
peripheral myelin protein 22
|
|
chr11_+_74950766
|
1.288
|
NM_001235
|
SERPINH1
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1)
|
|
chr8_+_1909437
|
1.271
|
NM_014867
|
KBTBD11
|
kelch repeat and BTB (POZ) domain containing 11
|
|
chr17_+_42641426
|
1.268
|
NM_001002841
|
MYL4
|
myosin, light chain 4, alkali; atrial, embryonic
|
|
chr10_+_99248605
|
1.265
|
NM_024954
|
UBTD1
|
ubiquitin domain containing 1
|
|
chr1_-_40140126
|
1.262
|
NM_001033081
NM_001033082
NM_005376
|
MYCL1
|
v-myc myelocytomatosis viral oncogene homolog 1, lung carcinoma derived (avian)
|
|
chr1_-_201411533
|
1.257
|
NM_004997
|
MYBPH
|
myosin binding protein H
|
|
chr7_-_37922902
|
1.248
|
|
SFRP4
|
secreted frizzled-related protein 4
|
|
chr9_-_139060285
|
1.245
|
|
NPDC1
|
neural proliferation, differentiation and control, 1
|
|
chr9_-_139060377
|
1.238
|
|
NPDC1
|
neural proliferation, differentiation and control, 1
|
|
chr2_+_33213111
|
1.238
|
NM_000627
NM_001166264
NM_001166265
NM_001166266
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chr3_+_35656012
|
1.222
|
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr2_-_161058120
|
1.203
|
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr1_-_201321711
|
1.186
|
|
MYOG
|
myogenin (myogenic factor 4)
|
|
chr10_-_62373828
|
1.183
|
NM_014836
|
RHOBTB1
|
Rho-related BTB domain containing 1
|
|
chrX_-_6156655
|
1.179
|
NM_181332
|
NLGN4X
|
neuroligin 4, X-linked
|
|
chr2_-_161058550
|
1.173
|
NM_002897
NM_016836
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr3_+_49034494
|
1.169
|
|
NDUFAF3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 3
|
|
chr7_+_100302885
|
1.167
|
NM_003302
|
TRIP6
|
thyroid hormone receptor interactor 6
|
|
chr7_+_115926855
|
1.165
|
|
CAV2
|
caveolin 2
|
|
chr11_+_113815317
|
1.165
|
NM_015523
|
REXO2
|
REX2, RNA exonuclease 2 homolog (S. cerevisiae)
|
|
chr22_-_17891777
|
1.163
|
|
CLDN5
|
claudin 5
|
|
chr5_+_95092384
|
1.160
|
NM_014899
|
RHOBTB3
|
Rho-related BTB domain containing 3
|
|
chrX_-_106130098
|
1.152
|
NM_001085354
NM_024657
|
MORC4
|
MORC family CW-type zinc finger 4
|
|
chr3_+_106568334
|
1.146
|
NM_001627
|
ALCAM
|
activated leukocyte cell adhesion molecule
|
|
chr16_+_7322751
|
1.146
|
NM_145891
NM_145892
NM_145893
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr1_-_143643324
|
1.142
|
NM_001002811
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr9_+_34980266
|
1.139
|
NM_001135005
|
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5
|
|
chr11_-_61415429
|
1.139
|
NM_021727
|
FADS3
|
fatty acid desaturase 3
|
|
chr16_+_54070302
|
1.137
|
NM_004530
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr17_-_74432800
|
1.133
|
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr8_+_22492198
|
1.127
|
NM_021630
|
PDLIM2
|
PDZ and LIM domain 2 (mystique)
|
|
chr7_-_150576660
|
1.126
|
NM_001003801
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3
|
|
chr12_-_54517241
|
1.121
|
|
MMP19
|
matrix metallopeptidase 19
|
|
chr6_-_46401363
|
1.110
|
NM_005822
|
RCAN2
|
regulator of calcineurin 2
|
|
chrX_+_99786044
|
1.109
|
|
SRPX2
|
sushi-repeat containing protein, X-linked 2
|
|
chr1_-_146398501
|
1.106
|
|
LOC100286793
FLJ39739
|
hypothetical LOC100286793
hypothetical FLJ39739
|
|
chr5_-_88235624
|
1.095
|
NM_001131005
NM_001193347
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chrX_+_99785818
|
1.094
|
NM_014467
|
SRPX2
|
sushi-repeat containing protein, X-linked 2
|
|
chr12_+_6703686
|
1.093
|
NM_001164093
|
COPS7A
|
COP9 constitutive photomorphogenic homolog subunit 7A (Arabidopsis)
|
|
chr17_-_74433048
|
1.087
|
NM_003255
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr14_-_22891263
|
1.086
|
NM_020372
|
SLC22A17
|
solute carrier family 22, member 17
|
|
chr17_-_45619038
|
1.085
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr22_+_36401558
|
1.082
|
NM_002305
|
LGALS1
|
lectin, galactoside-binding, soluble, 1
|
|
chr11_-_63770983
|
1.081
|
NM_138689
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B
|
|
chr9_+_34980715
|
1.078
|
|
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5
|
|
chr8_+_98857190
|
1.075
|
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta
|
|
chr17_-_37221671
|
1.071
|
|
LEPREL4
|
leprecan-like 4
|