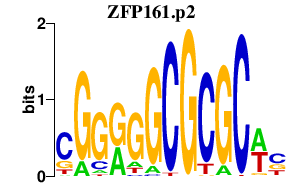
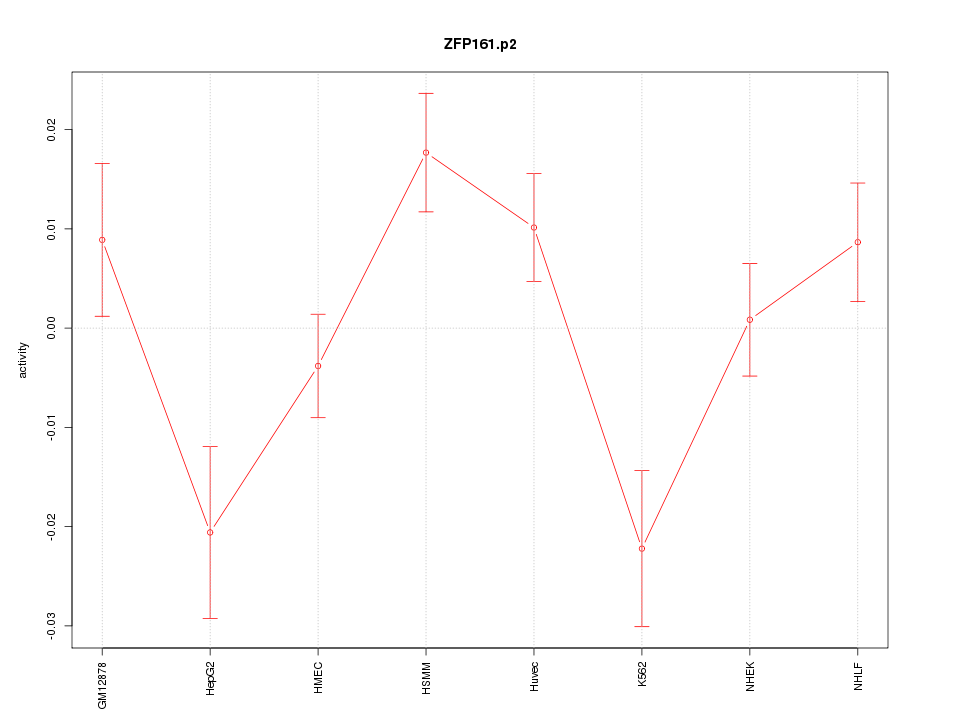
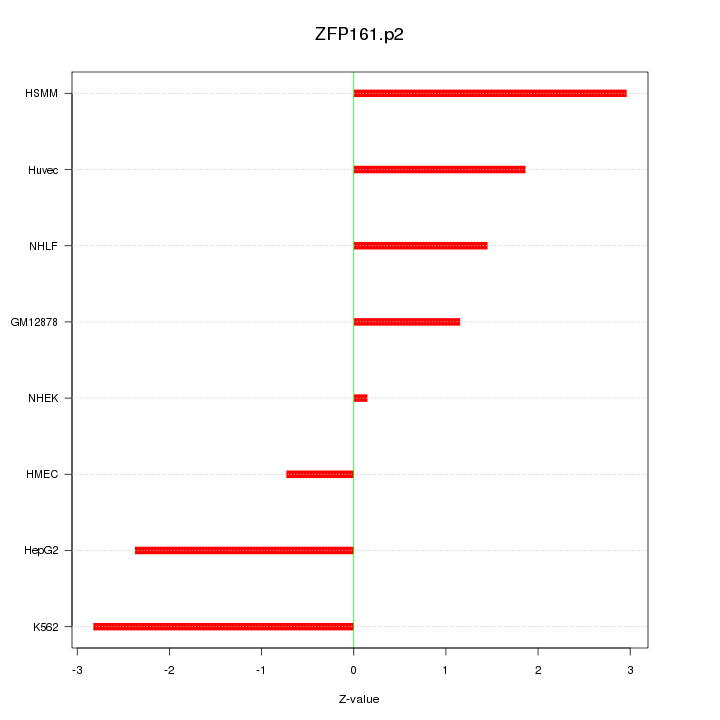
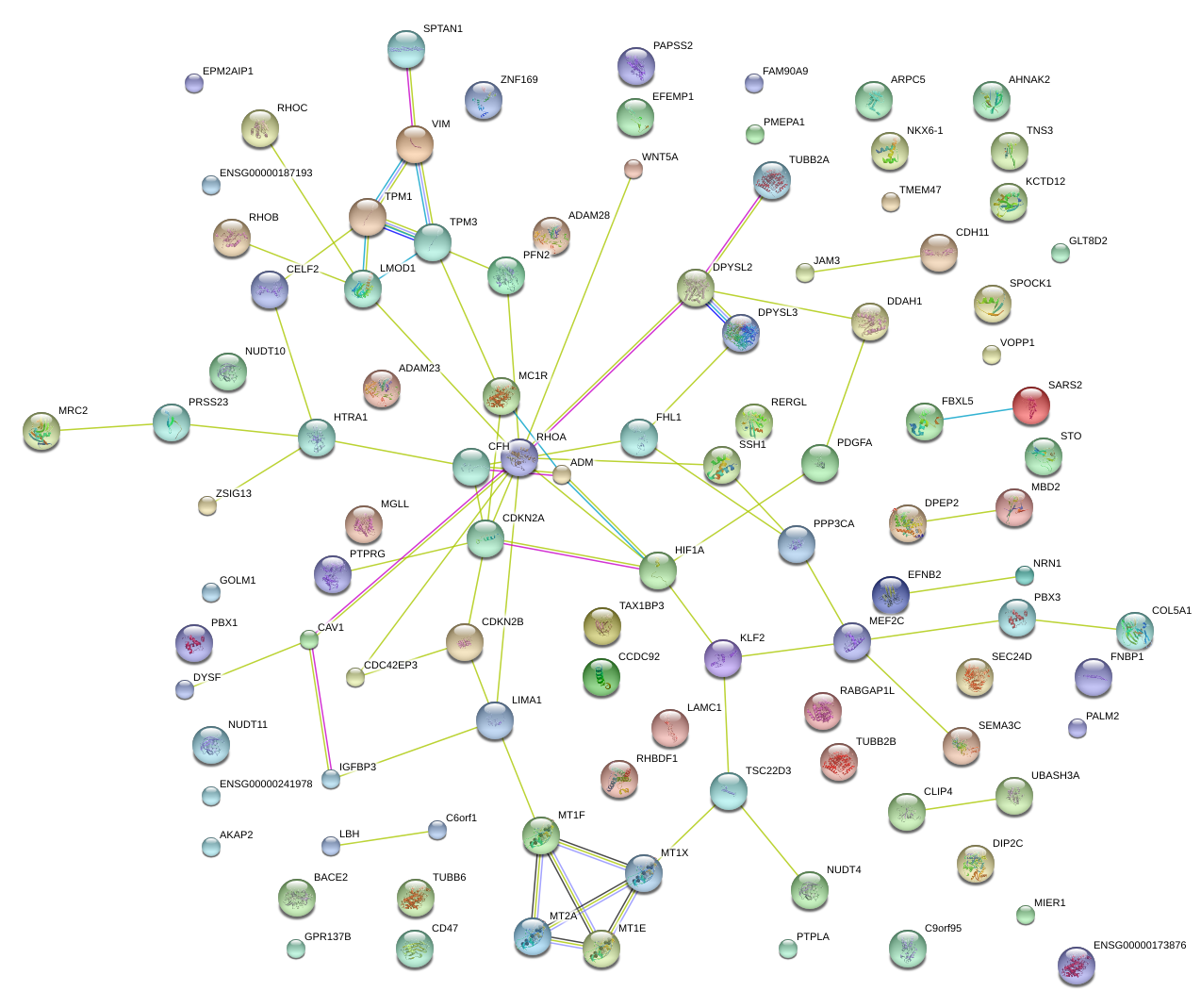
|
chr1_+_234372454
|
4.183
|
NM_003272
|
GPR137B
|
G protein-coupled receptor 137B
|
|
chr10_+_124210980
|
3.606
|
NM_002775
|
HTRA1
|
HtrA serine peptidase 1
|
|
chr10_+_124211353
|
3.397
|
|
HTRA1
|
HtrA serine peptidase 1
|
|
chr3_-_129024665
|
3.099
|
NM_001003794
|
MGLL
|
monoglyceride lipase
|
|
chr3_-_151171190
|
2.579
|
NM_002628
NM_053024
|
PFN2
|
profilin 2
|
|
chr9_-_21984379
|
2.393
|
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4)
|
|
chr11_+_86189138
|
2.142
|
NM_007173
|
PRSS23
|
protease, serine, 23
|
|
chr3_-_55496607
|
2.132
|
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr14_-_104515738
|
2.083
|
NM_138420
|
AHNAK2
|
AHNAK nucleoprotein 2
|
|
chrX_-_34585287
|
1.886
|
NM_031442
|
TMEM47
|
transmembrane protein 47
|
|
chr9_-_21984414
|
1.885
|
NM_058195
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4)
|
|
chr13_-_76358433
|
1.829
|
NM_138444
|
KCTD12
|
potassium channel tetramerisation domain containing 12
|
|
chr10_-_725522
|
1.816
|
NM_014974
|
DIP2C
|
DIP2 disco-interacting protein 2 homolog C (Drosophila)
|
|
chr3_-_109292444
|
1.791
|
NM_001777
NM_198793
|
CD47
|
CD47 molecule
|
|
chr3_-_129023850
|
1.653
|
NM_007283
|
MGLL
|
monoglyceride lipase
|
|
chr10_+_11100111
|
1.623
|
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr3_-_55496370
|
1.611
|
NM_003392
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr10_+_11099860
|
1.566
|
NM_006561
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr9_-_21984329
|
1.518
|
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4)
|
|
chr1_+_172395277
|
1.499
|
|
RABGAP1L
|
RAB GTPase activating protein 1-like
|
|
chr1_+_67168470
|
1.459
|
NM_001077701
NM_001077704
|
MIER1
|
mesoderm induction early response 1 homolog (Xenopus laevis)
|
|
chr1_+_172395170
|
1.448
|
NM_014857
|
RABGAP1L
|
RAB GTPase activating protein 1-like
|
|
chrX_-_51256166
|
1.382
|
NM_018159
|
NUDT11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11
|
|
chrX_+_135057202
|
1.335
|
NM_001159702
NM_001159703
NM_001449
|
FHL1
|
four and a half LIM domains 1
|
|
chr10_+_11100075
|
1.330
|
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr2_+_30307900
|
1.254
|
NM_030915
|
LBH
|
limb bud and heart development homolog (mouse)
|
|
chr15_+_61127367
|
1.241
|
NM_001018008
|
TPM1
|
tropomyosin 1 (alpha)
|
|
chr2_-_37752272
|
1.222
|
|
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3
|
|
chr9_-_76892860
|
1.215
|
NM_001127603
NM_017881
|
C9orf95
|
chromosome 9 open reading frame 95
|
|
chrX_-_51256035
|
1.167
|
|
NUDT11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11
|
|
chr10_+_17310475
|
1.162
|
|
VIM
|
vimentin
|
|
chr7_-_80386602
|
1.138
|
NM_006379
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C
|
|
chr7_-_80386258
|
1.133
|
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C
|
|
chr2_-_56004366
|
1.127
|
NM_001039348
NM_001039349
|
EFEMP1
|
EGF containing fibulin-like extracellular matrix protein 1
|
|
chr9_-_87904306
|
1.123
|
NM_016548
|
GOLM1
|
golgi membrane protein 1
|
|
chr9_-_87904280
|
1.105
|
|
GOLM1
|
golgi membrane protein 1
|
|
chr4_-_102487356
|
1.097
|
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme
|
|
chr2_+_207016525
|
1.092
|
NM_003812
|
ADAM23
|
ADAM metallopeptidase domain 23
|
|
chr7_-_55607564
|
1.081
|
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1
|
|
chr9_-_131845277
|
1.077
|
NM_015033
|
FNBP1
|
formin binding protein 1
|
|
chr9_-_87904215
|
1.066
|
|
GOLM1
|
golgi membrane protein 1
|
|
chr11_+_133444029
|
1.065
|
NM_032801
|
JAM3
|
junctional adhesion molecule 3
|
|
chr7_-_55607625
|
1.064
|
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1
|
|
chr12_-_107775462
|
1.063
|
NM_001161330
NM_018984
|
SSH1
|
slingshot homolog 1 (Drosophila)
|
|
chr19_+_16296632
|
1.032
|
NM_016270
|
KLF2
|
Kruppel-like factor 2 (lung)
|
|
chr10_-_17699371
|
1.028
|
NM_014241
|
PTPLA
|
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A
|
|
chr1_-_85703321
|
1.025
|
NM_012137
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr7_-_55607598
|
1.016
|
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1
|
|
chr2_+_71547325
|
1.011
|
NM_001130455
NM_001130982
NM_001130983
NM_001130984
NM_001130985
NM_001130986
NM_001130987
|
DYSF
|
dysferlin, limb girdle muscular dystrophy 2B (autosomal recessive)
|
|
chr11_+_133444175
|
1.009
|
|
JAM3
|
junctional adhesion molecule 3
|
|
chr5_-_146869811
|
0.989
|
NM_001197294
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr17_+_58058783
|
0.979
|
|
MRC2
|
mannose receptor, C type 2
|
|
chr18_-_50004770
|
0.978
|
|
MBD2
|
methyl-CpG binding domain protein 2
|
|
chr5_-_88215034
|
0.977
|
NM_001193350
NM_002397
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr7_-_55607692
|
0.972
|
NM_030796
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1
|
|
chr12_-_123023052
|
0.972
|
NM_025140
|
CCDC92
|
coiled-coil domain containing 92
|
|
chr17_+_58058479
|
0.956
|
NM_006039
|
MRC2
|
mannose receptor, C type 2
|
|
chr9_+_127549437
|
0.952
|
NM_006195
|
PBX3
|
pre-B-cell leukemia homeobox 3
|
|
chr20_-_55718351
|
0.946
|
NM_020182
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr17_+_58058774
|
0.942
|
|
MRC2
|
mannose receptor, C type 2
|
|
chr8_+_26491317
|
0.936
|
NM_001386
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr6_-_5952631
|
0.930
|
NM_016588
|
NRN1
|
neuritin 1
|
|
chr13_-_105985313
|
0.917
|
NM_004093
|
EFNB2
|
ephrin-B2
|
|
chr9_+_136673464
|
0.914
|
NM_000093
|
COL5A1
|
collagen, type V, alpha 1
|
|
chr4_-_119976631
|
0.894
|
|
SEC24D
|
SEC24 family, member D (S. cerevisiae)
|
|
chr10_+_89409332
|
0.894
|
NM_001015880
NM_004670
|
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2
|
|
chr7_-_47588266
|
0.889
|
|
TNS3
|
tensin 3
|
|
chr12_-_48963505
|
0.888
|
NM_001113546
NM_016357
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr1_+_181259317
|
0.886
|
|
|
|
|
chrX_-_106905657
|
0.880
|
NM_198057
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr11_+_10283171
|
0.877
|
NM_001124
|
ADM
|
adrenomedullin
|
|
chr16_-_62573
|
0.872
|
NM_022450
|
RHBDF1
|
rhomboid 5 homolog 1 (Drosophila)
|
|
chr3_+_61522219
|
0.870
|
NM_002841
|
PTPRG
|
protein tyrosine phosphatase, receptor type, G
|
|
chr21_+_41461962
|
0.850
|
|
BACE2
|
beta-site APP-cleaving enzyme 2
|
|
chr1_+_181258952
|
0.849
|
NM_002293
|
LAMC1
|
laminin, gamma 1 (formerly LAMB2)
|
|
chr16_-_63713382
|
0.848
|
NM_001797
|
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast)
|
|
chr9_+_111582317
|
0.844
|
NM_053016
NM_007203
NM_147150
|
PALM2
PALM2-AKAP2
|
paralemmin 2
PALM2-AKAP2 readthrough
|
|
chr5_+_176493233
|
0.844
|
NM_022455
|
NSD1
|
nuclear receptor binding SET domain protein 1
|
|
chr3_-_49424404
|
0.843
|
|
RHOA
|
ras homolog gene family, member A
|
|
chr5_-_136862786
|
0.841
|
NM_004598
|
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1
|
|
chr7_-_525556
|
0.839
|
|
PDGFA
|
platelet-derived growth factor alpha polypeptide
|
|
chr4_-_15266042
|
0.835
|
|
FBXL5
|
F-box and leucine-rich repeat protein 5
|
|
chr4_-_15266072
|
0.829
|
NM_001193534
NM_001193535
NM_012161
|
FBXL5
|
F-box and leucine-rich repeat protein 5
|
|
chr5_-_88214720
|
0.829
|
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr3_-_49424413
|
0.827
|
|
RHOA
|
ras homolog gene family, member A
|
|
chr1_-_85703198
|
0.826
|
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr18_+_12298236
|
0.825
|
|
TUBB6
|
tubulin, beta 6
|
|
chr3_-_49424397
|
0.822
|
|
RHOA
|
ras homolog gene family, member A
|
|
chr7_-_45927263
|
0.816
|
NM_000598
NM_001013398
|
IGFBP3
|
insulin-like growth factor binding protein 3
|
|
chr3_-_49424427
|
0.813
|
|
RHOA
|
ras homolog gene family, member A
|
|
chr3_-_49424506
|
0.812
|
|
RHOA
|
ras homolog gene family, member A
|
|
chr3_-_49424361
|
0.808
|
|
RHOA
|
ras homolog gene family, member A
|
|
chr3_-_49424529
|
0.804
|
NM_001664
|
RHOA
|
ras homolog gene family, member A
|
|
chr3_-_49424380
|
0.801
|
|
RHOA
|
ras homolog gene family, member A
|
|
chr4_-_15265993
|
0.799
|
|
FBXL5
|
F-box and leucine-rich repeat protein 5
|
|
chr3_-_49424322
|
0.788
|
|
RHOA
|
ras homolog gene family, member A
|
|
chr17_-_3518628
|
0.783
|
|
TAX1BP3
|
Tax1 (human T-cell leukemia virus type I) binding protein 3
|
|
chr1_-_181871447
|
0.779
|
|
ARPC5
|
actin related protein 2/3 complex, subunit 5, 16kDa
|
|
chrX_-_106905545
|
0.775
|
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr14_+_61231795
|
0.772
|
NM_001530
NM_181054
|
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor)
|
|
chr9_+_130354490
|
0.767
|
NM_001130438
NM_001195532
NM_003127
|
SPTAN1
|
spectrin, alpha, non-erythrocytic 1 (alpha-fodrin)
|
|
chr7_+_115952325
|
0.764
|
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr9_-_87904844
|
0.758
|
NM_177937
|
GOLM1
|
golgi membrane protein 1
|
|
chr16_+_55199978
|
0.756
|
NM_005953
|
MT2A
|
metallothionein 2A
|
|
chr2_+_29191711
|
0.750
|
NM_024692
|
CLIP4
|
CAP-GLY domain containing linker protein family, member 4
|
|
chr15_+_46957556
|
0.749
|
NM_014335
|
EID1
|
EP300 interacting inhibitor of differentiation 1
|
|
chr1_-_181871416
|
0.745
|
|
ARPC5
|
actin related protein 2/3 complex, subunit 5, 16kDa
|
|
chr7_-_525336
|
0.741
|
|
PDGFA
|
platelet-derived growth factor alpha polypeptide
|
|
chr17_-_3518568
|
0.728
|
|
TAX1BP3
|
Tax1 (human T-cell leukemia virus type I) binding protein 3
|
|
chr17_-_3518654
|
0.716
|
NM_014604
|
TAX1BP3
|
Tax1 (human T-cell leukemia virus type I) binding protein 3
|
|
chr8_-_67687980
|
0.711
|
NM_001080416
NM_001144755
|
MYBL1
LOC645895
|
v-myb myeloblastosis viral oncogene homolog (avian)-like 1
hypothetical LOC645895
|
|
chr14_+_22411200
|
0.711
|
|
LRP10
|
low density lipoprotein receptor-related protein 10
|
|
chr14_+_66777578
|
0.708
|
NM_022474
|
MPP5
|
membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5)
|
|
chr9_-_16860664
|
0.699
|
|
BNC2
|
basonuclin 2
|
|
chr17_-_1341599
|
0.696
|
NM_033375
|
MYO1C
|
myosin IC
|
|
chr4_-_102487649
|
0.695
|
NM_000944
NM_001130691
NM_001130692
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme
|
|
chr11_-_6396864
|
0.695
|
NM_001164
|
APBB1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65)
|
|
chr10_-_17699309
|
0.691
|
|
PTPLA
|
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A
|
|
chr7_+_156624424
|
0.688
|
|
UBE3C
|
ubiquitin protein ligase E3C
|
|
chr9_-_16860719
|
0.687
|
NM_017637
|
BNC2
|
basonuclin 2
|
|
chr5_-_171547700
|
0.687
|
|
STK10
|
serine/threonine kinase 10
|
|
chr7_+_79602023
|
0.686
|
NM_002069
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1
|
|
chr14_+_64949447
|
0.681
|
|
FUT8
|
fucosyltransferase 8 (alpha (1,6) fucosyltransferase)
|
|
chr6_-_166960690
|
0.678
|
NM_021135
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2
|
|
chr1_-_181871462
|
0.677
|
|
ARPC5
|
actin related protein 2/3 complex, subunit 5, 16kDa
|
|
chr4_-_119976736
|
0.676
|
NM_014822
|
SEC24D
|
SEC24 family, member D (S. cerevisiae)
|
|
chr19_+_16296734
|
0.674
|
|
KLF2
|
Kruppel-like factor 2 (lung)
|
|
chr13_+_113546943
|
0.667
|
|
GAS6
|
growth arrest-specific 6
|
|
chr7_-_55607510
|
0.667
|
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1
|
|
chr1_-_181871490
|
0.667
|
|
ARPC5
|
actin related protein 2/3 complex, subunit 5, 16kDa
|
|
chr5_+_176493488
|
0.663
|
|
NSD1
|
nuclear receptor binding SET domain protein 1
|
|
chr3_-_126257425
|
0.660
|
NM_020733
|
HEG1
|
HEG homolog 1 (zebrafish)
|
|
chr8_+_38973681
|
0.658
|
|
ADAM9
|
ADAM metallopeptidase domain 9
|
|
chr5_+_86599350
|
0.657
|
|
RASA1
|
RAS p21 protein activator (GTPase activating protein) 1
|
|
chr2_-_175255717
|
0.656
|
NM_001077269
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr4_-_54625522
|
0.653
|
|
CHIC2
|
cysteine-rich hydrophobic domain 2
|
|
chr1_-_181871508
|
0.649
|
NM_005717
|
ARPC5
|
actin related protein 2/3 complex, subunit 5, 16kDa
|
|
chr9_-_13269562
|
0.648
|
|
MPDZ
|
multiple PDZ domain protein
|
|
chr1_-_181871473
|
0.640
|
|
ARPC5
|
actin related protein 2/3 complex, subunit 5, 16kDa
|
|
chr18_+_9698269
|
0.640
|
|
RAB31
|
RAB31, member RAS oncogene family
|
|
chr3_-_38666122
|
0.639
|
NM_000335
NM_001099404
NM_001099405
NM_198056
NM_001160160
NM_001160161
|
SCN5A
|
sodium channel, voltage-gated, type V, alpha subunit
|
|
chr8_+_94998258
|
0.637
|
NM_018444
NM_001161780
NM_001161779
NM_001161781
|
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1
|
|
chr1_+_84316217
|
0.637
|
NM_002731
NM_207578
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr1_+_216586199
|
0.637
|
|
TGFB2
|
transforming growth factor, beta 2
|
|
chr7_-_150576957
|
0.636
|
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3
|
|
chr7_+_68701704
|
0.635
|
NM_001127231
NM_001127232
NM_015570
|
AUTS2
|
autism susceptibility candidate 2
|
|
chr16_-_63713466
|
0.635
|
|
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast)
|
|
chr4_-_5945596
|
0.634
|
NM_001014809
|
CRMP1
|
collapsin response mediator protein 1
|
|
chr4_-_54625463
|
0.632
|
|
CHIC2
|
cysteine-rich hydrophobic domain 2
|
|
chr2_-_161058120
|
0.625
|
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr1_+_156229686
|
0.625
|
NM_018240
|
KIRREL
|
kin of IRRE like (Drosophila)
|
|
chr22_+_28329560
|
0.625
|
|
NF2
|
neurofibromin 2 (merlin)
|
|
chr18_+_12298242
|
0.621
|
NM_032525
|
TUBB6
|
tubulin, beta 6
|
|
chr4_+_40953666
|
0.616
|
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chrX_-_17789276
|
0.610
|
NM_001172732
NM_001172739
NM_001172743
NM_021785
|
RAI2
|
retinoic acid induced 2
|
|
chr3_-_49370682
|
0.609
|
|
GPX1
|
glutathione peroxidase 1
|
|
chr2_+_241586927
|
0.605
|
NM_001080437
|
SNED1
|
sushi, nidogen and EGF-like domains 1
|
|
chr15_-_54073024
|
0.603
|
NM_006154
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr18_-_50004932
|
0.602
|
|
MBD2
|
methyl-CpG binding domain protein 2
|
|
chr14_-_91483757
|
0.600
|
NM_006329
|
FBLN5
|
fibulin 5
|
|
chr15_-_56829225
|
0.600
|
NM_001110
|
ADAM10
|
ADAM metallopeptidase domain 10
|
|
chr1_+_181871830
|
0.599
|
NM_015149
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr4_+_40953698
|
0.597
|
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr16_-_63713485
|
0.595
|
|
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast)
|
|
chr9_-_112840064
|
0.590
|
NM_001401
NM_057159
|
LPAR1
|
lysophosphatidic acid receptor 1
|
|
chr4_+_40953652
|
0.590
|
NM_004181
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr8_+_26491383
|
0.587
|
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr22_+_28329605
|
0.587
|
|
NF2
|
neurofibromin 2 (merlin)
|
|
chr14_+_60858151
|
0.586
|
NM_006255
|
PRKCH
|
protein kinase C, eta
|
|
chr1_-_103346639
|
0.586
|
NM_001190709
NM_001854
NM_080629
NM_080630
|
COL11A1
|
collagen, type XI, alpha 1
|
|
chr12_-_123023312
|
0.583
|
|
CCDC92
|
coiled-coil domain containing 92
|
|
chr3_+_49424757
|
0.579
|
|
TCTA
|
T-cell leukemia translocation altered gene
|
|
chr10_+_3099708
|
0.576
|
NM_002627
|
PFKP
|
phosphofructokinase, platelet
|
|
chr5_+_17270474
|
0.576
|
NM_006317
|
BASP1
|
brain abundant, membrane attached signal protein 1
|
|
chr1_+_203064397
|
0.570
|
NM_001005388
NM_001005389
NM_001160332
NM_001160333
NM_015090
|
NFASC
|
neurofascin
|
|
chr14_-_29466562
|
0.569
|
NM_002742
|
PRKD1
|
protein kinase D1
|
|
chr20_+_8997660
|
0.566
|
NM_001172646
|
PLCB4
|
phospholipase C, beta 4
|
|
chr21_-_27261276
|
0.562
|
NM_007038
|
ADAMTS5
|
ADAM metallopeptidase with thrombospondin type 1 motif, 5
|
|
chr11_+_17697685
|
0.557
|
NM_002478
|
MYOD1
|
myogenic differentiation 1
|
|
chr5_-_171547844
|
0.556
|
|
STK10
|
serine/threonine kinase 10
|
|
chr3_-_125085834
|
0.555
|
NM_053025
NM_053026
NM_053027
NM_053028
|
MYLK
|
myosin light chain kinase
|
|
chr2_+_27967102
|
0.554
|
|
BRE
|
brain and reproductive organ-expressed (TNFRSF1A modulator)
|
|
chr11_+_65594361
|
0.553
|
NM_018026
|
PACS1
|
phosphofurin acidic cluster sorting protein 1
|
|
chr14_-_91483325
|
0.549
|
|
FBLN5
|
fibulin 5
|
|
chr16_+_57055494
|
0.547
|
NM_001130487
|
NDRG4
|
NDRG family member 4
|
|
chr5_-_111121691
|
0.542
|
NM_001142477
NM_001142476
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr12_-_45759691
|
0.541
|
NM_001143668
NM_181847
|
AMIGO2
|
adhesion molecule with Ig-like domain 2
|
|
chr5_+_135392482
|
0.540
|
NM_000358
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa
|
|
chr1_+_222437537
|
0.537
|
NM_003676
|
DEGS1
|
degenerative spermatocyte homolog 1, lipid desaturase (Drosophila)
|
|
chr7_-_47588652
|
0.536
|
|
TNS3
|
tensin 3
|
|
chr19_-_4351470
|
0.536
|
NM_003025
|
SH3GL1
|
SH3-domain GRB2-like 1
|
|
chr1_+_148388719
|
0.535
|
NM_016274
|
PLEKHO1
|
pleckstrin homology domain containing, family O member 1
|
|
chr12_-_31635089
|
0.533
|
NM_144973
|
DENND5B
|
DENN/MADD domain containing 5B
|
|
chr15_+_78858763
|
0.530
|
NM_018689
|
KIAA1199
|
KIAA1199
|
|
chr9_+_76893209
|
0.523
|
NM_012383
|
OSTF1
|
osteoclast stimulating factor 1
|
|
chr3_+_185375379
|
0.520
|
|
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit
|
|
chr10_+_90740364
|
0.520
|
|
FAS
|
Fas (TNF receptor superfamily, member 6)
|
|
chr16_+_57055129
|
0.516
|
|
NDRG4
|
NDRG family member 4
|
|
chr5_+_119827903
|
0.515
|
NM_016644
|
PRR16
|
proline rich 16
|
|
chr15_-_32118477
|
0.515
|
NM_020371
|
AVEN
|
apoptosis, caspase activation inhibitor
|