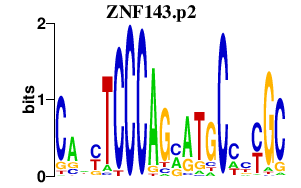
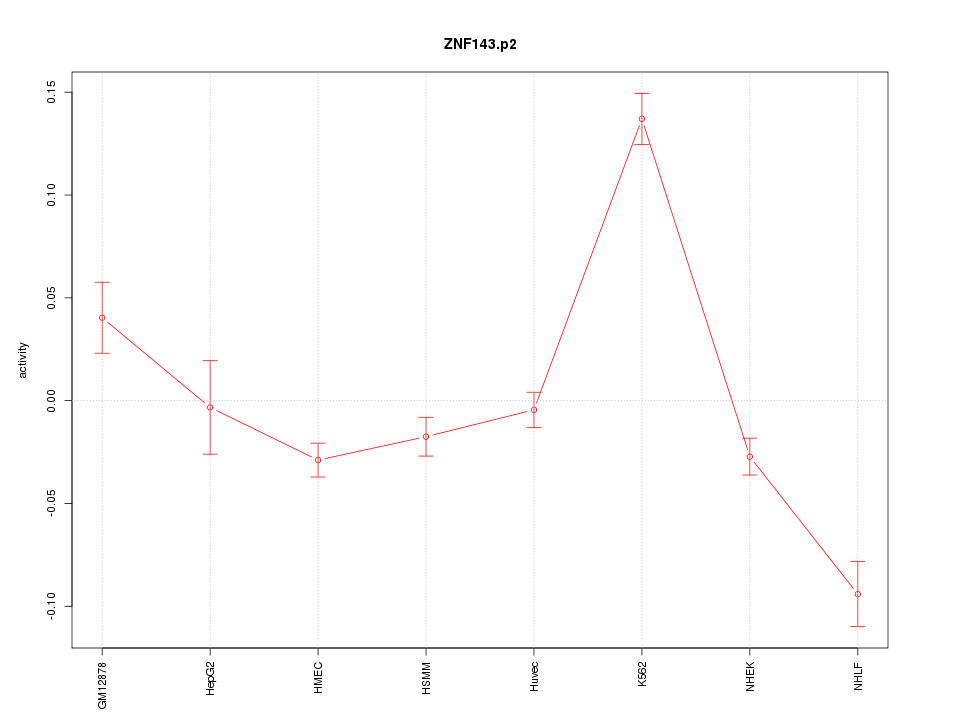
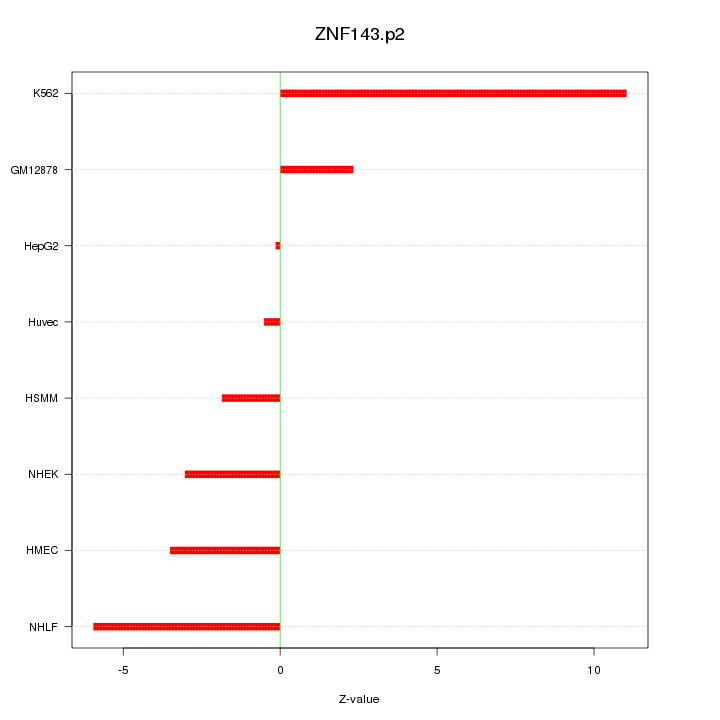
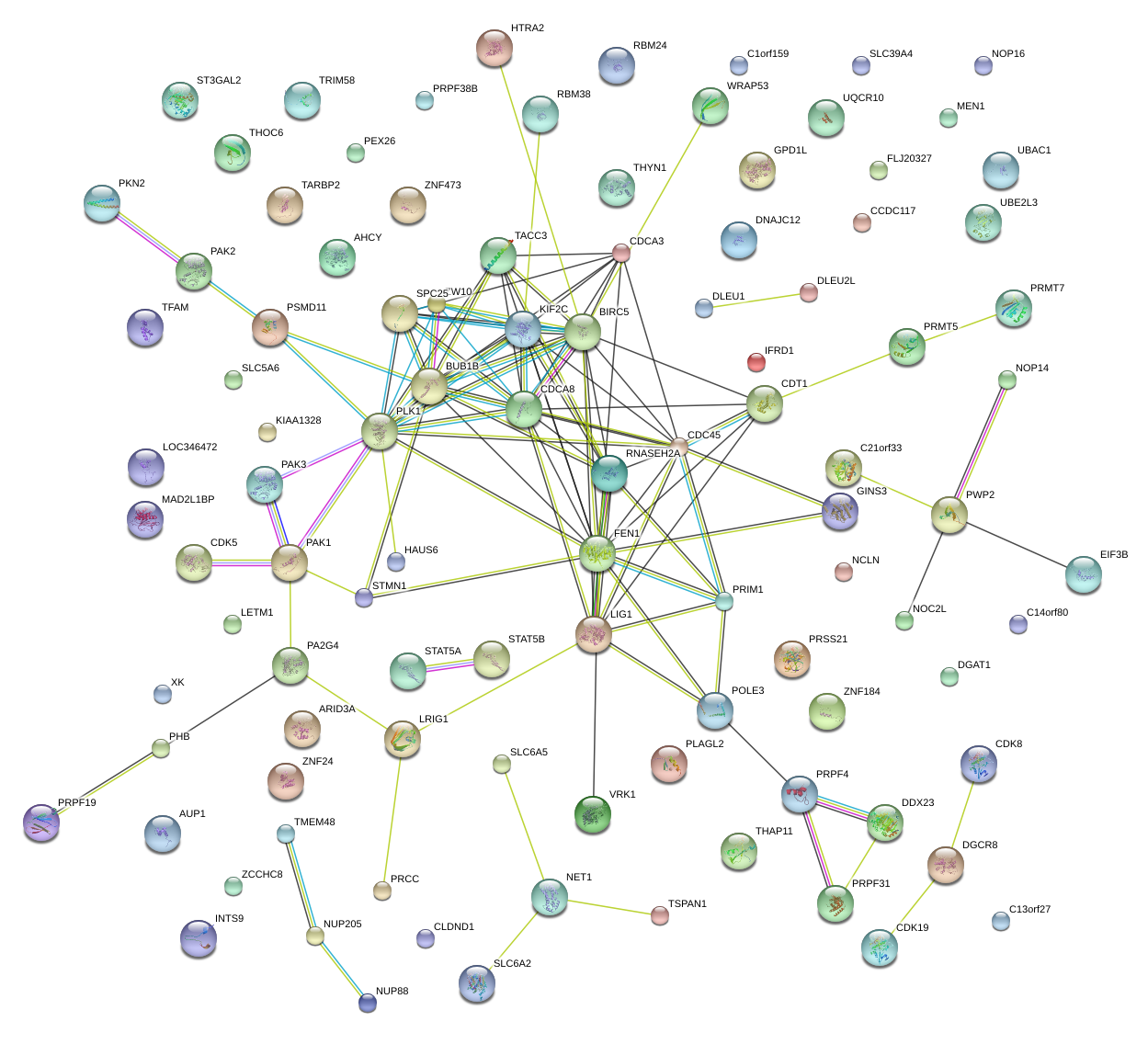
|
chr19_-_53365371
|
6.916
|
NM_000234
|
LIG1
|
ligase I, DNA, ATP-dependent
|
|
chr1_-_54076521
|
5.358
|
|
TMEM48
|
transmembrane protein 48
|
|
chr2_+_74610635
|
5.331
|
|
HTRA2
|
HtrA serine peptidase 2
|
|
chr1_-_54076695
|
5.221
|
NM_001168551
NM_018087
|
TMEM48
|
transmembrane protein 48
|
|
chr1_+_44978145
|
5.074
|
|
KIF2C
|
kinesin family member 2C
|
|
chr2_+_74610666
|
4.684
|
|
HTRA2
|
HtrA serine peptidase 2
|
|
chr4_-_2934945
|
4.619
|
|
NOP14
|
NOP14 nucleolar protein homolog (yeast)
|
|
chr2_+_74610395
|
4.553
|
|
HTRA2
|
HtrA serine peptidase 2
|
|
chr2_+_74610494
|
4.451
|
|
HTRA2
|
HtrA serine peptidase 2
|
|
chr2_+_74610624
|
4.407
|
|
HTRA2
|
HtrA serine peptidase 2
|
|
chr1_+_44978120
|
4.251
|
|
KIF2C
|
kinesin family member 2C
|
|
chr12_+_54784625
|
4.139
|
|
PA2G4
|
proliferation-associated 2G4, 38kDa
|
|
chr20_+_55399847
|
3.948
|
NM_017495
NM_183425
|
RBM38
|
RNA binding motif protein 38
|
|
chr12_+_54784369
|
3.893
|
NM_006191
|
PA2G4
|
proliferation-associated 2G4, 38kDa
|
|
chr22_+_27498679
|
3.857
|
|
CCDC117
|
coiled-coil domain containing 117
|
|
chr2_+_74610751
|
3.847
|
|
HTRA2
|
HtrA serine peptidase 2
|
|
chr8_-_28803616
|
3.761
|
NM_001145159
NM_018250
|
INTS9
|
integrator complex subunit 9
|
|
chr1_+_44978053
|
3.664
|
NM_006845
|
KIF2C
|
kinesin family member 2C
|
|
chr12_+_54784632
|
3.574
|
|
PA2G4
|
proliferation-associated 2G4, 38kDa
|
|
chr7_-_150385912
|
3.464
|
NM_001164410
NM_004935
|
CDK5
|
cyclin-dependent kinase 5
|
|
chr12_+_54784600
|
3.440
|
|
PA2G4
|
proliferation-associated 2G4, 38kDa
|
|
chr8_-_28803341
|
3.437
|
NM_001172562
|
INTS9
|
integrator complex subunit 9
|
|
chr7_-_150385867
|
3.409
|
|
CDK5
|
cyclin-dependent kinase 5
|
|
chr12_+_54784681
|
3.397
|
|
PA2G4
|
proliferation-associated 2G4, 38kDa
|
|
chr2_+_74610333
|
3.371
|
|
HTRA2
|
HtrA serine peptidase 2
|
|
chr17_+_73721924
|
3.360
|
|
BIRC5
|
baculoviral IAP repeat containing 5
|
|
chr17_+_73721871
|
3.343
|
NM_001012270
NM_001012271
NM_001168
|
BIRC5
|
baculoviral IAP repeat containing 5
|
|
chr17_-_44847214
|
3.329
|
|
PHB
|
prohibitin
|
|
chr17_-_44847215
|
3.257
|
NM_002634
|
PHB
|
prohibitin
|
|
chr9_-_137992967
|
3.221
|
NM_016172
|
UBAC1
|
UBA domain containing 1
|
|
chr9_-_137992766
|
3.061
|
|
UBAC1
|
UBA domain containing 1
|
|
chr19_+_59310601
|
2.993
|
NM_015629
|
PRPF31
|
PRP31 pre-mRNA processing factor 31 homolog (S. cerevisiae)
|
|
chr6_-_27548438
|
2.955
|
NM_007149
|
ZNF184
|
zinc finger protein 184
|
|
chr14_+_96333434
|
2.954
|
NM_003384
|
VRK1
|
vaccinia related kinase 1
|
|
chr4_+_1693031
|
2.929
|
NM_006342
|
TACC3
|
transforming, acidic coiled-coil containing protein 3
|
|
chr13_-_49554089
|
2.897
|
|
DLEU2
|
deleted in lymphocytic leukemia 2 (non-protein coding)
|
|
chr6_+_43705254
|
2.856
|
NM_001003690
|
MAD2L1BP
|
MAD2L1 binding protein
|
|
chr14_-_22468393
|
2.761
|
|
PRMT5
|
protein arginine methyltransferase 5
|
|
chr12_+_54784591
|
2.741
|
|
PA2G4
|
proliferation-associated 2G4, 38kDa
|
|
chr12_+_54784614
|
2.668
|
|
PA2G4
|
proliferation-associated 2G4, 38kDa
|
|
chrX_+_37429927
|
2.611
|
NM_021083
|
XK
|
X-linked Kx blood group (McLeod syndrome)
|
|
chr14_-_22468445
|
2.537
|
NM_001039619
NM_006109
|
PRMT5
|
protein arginine methyltransferase 5
|
|
chr19_+_12778391
|
2.524
|
|
RNASEH2A
|
ribonuclease H2, subunit A
|
|
chr15_+_38240501
|
2.474
|
NM_001211
|
BUB1B
|
budding uninhibited by benzimidazoles 1 homolog beta (yeast)
|
|
chr4_-_1827475
|
2.462
|
NM_012318
|
LETM1
|
leucine zipper-EF-hand containing transmembrane protein 1
|
|
chr9_-_115212479
|
2.450
|
|
POLE3
|
polymerase (DNA directed), epsilon 3 (p17 subunit)
|
|
chr22_+_17847467
|
2.408
|
|
CDC45
|
cell division cycle 45 homolog (S. cerevisiae)
|
|
chr4_-_2934909
|
2.349
|
NM_003703
|
NOP14
|
NOP14 nucleolar protein homolog (yeast)
|
|
chr15_+_38240555
|
2.336
|
|
BUB1B
|
budding uninhibited by benzimidazoles 1 homolog beta (yeast)
|
|
chr13_+_25726238
|
2.291
|
|
CDK8
|
cyclin-dependent kinase 8
|
|
chr16_-_69030421
|
2.274
|
NM_006927
|
ST3GAL2
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 2
|
|
chr11_-_60430615
|
2.271
|
NM_014502
|
PRPF19
|
PRP19/PSO4 pre-mRNA processing factor 19 homolog (S. cerevisiae)
|
|
chr13_+_49554307
|
2.256
|
|
DLEU1
|
deleted in lymphocytic leukemia 1 (non-protein coding)
|
|
chr2_-_74610323
|
2.246
|
|
AUP1
|
ancient ubiquitous protein 1
|
|
chr2_+_74610039
|
2.244
|
NM_013247
NM_145074
|
HTRA2
|
HtrA serine peptidase 2
|
|
chr16_+_2807228
|
2.219
|
|
PRSS21
|
protease, serine, 21 (testisin)
|
|
chr7_+_111877699
|
2.216
|
NM_001550
|
IFRD1
|
interferon-related developmental regulator 1
|
|
chr19_+_55221106
|
2.214
|
|
ZNF473
|
zinc finger protein 473
|
|
chr1_+_37930716
|
2.212
|
NM_018101
|
CDCA8
|
cell division cycle associated 8
|
|
chr19_+_55221088
|
2.128
|
|
ZNF473
|
zinc finger protein 473
|
|
chr2_-_74610331
|
2.124
|
|
AUP1
|
ancient ubiquitous protein 1
|
|
chr5_+_5475766
|
2.103
|
|
|
|
|
chr1_-_1041321
|
2.100
|
|
C1orf159
|
chromosome 1 open reading frame 159
|
|
chr10_-_69267774
|
2.064
|
NM_021800
NM_201262
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12
|
|
chr2_-_74610481
|
2.061
|
NM_181575
|
AUP1
|
ancient ubiquitous protein 1
|
|
chr12_-_121551424
|
2.046
|
NM_017612
|
ZCCHC8
|
zinc finger, CCHC domain containing 8
|
|
chr20_-_30259007
|
2.034
|
NM_002657
|
PLAGL2
|
pleiomorphic adenoma gene-like 2
|
|
chr4_-_2934889
|
2.026
|
|
NOP14
|
NOP14 nucleolar protein homolog (yeast)
|
|
chr18_-_31178415
|
2.017
|
NM_006965
|
ZNF24
|
zinc finger protein 24
|
|
chr19_+_12778409
|
1.998
|
NM_006397
|
RNASEH2A
|
ribonuclease H2, subunit A
|
|
chr2_-_74610332
|
1.988
|
|
AUP1
|
ancient ubiquitous protein 1
|
|
chr12_-_55432348
|
1.980
|
NM_000946
|
PRIM1
|
primase, DNA, polypeptide 1 (49kDa)
|
|
chr16_+_66902369
|
1.967
|
NM_001184824
NM_019023
|
PRMT7
|
protein arginine methyltransferase 7
|
|
chr22_+_16940755
|
1.946
|
|
PEX26
|
peroxisomal biogenesis factor 26
|
|
chr11_-_64334752
|
1.942
|
NM_000244
NM_130799
NM_130802
|
MEN1
|
multiple endocrine neoplasia I
|
|
chr12_+_52181643
|
1.925
|
NM_134324
|
TARBP2
|
TAR (HIV-1) RNA binding protein 2
|
|
chr12_-_47532171
|
1.909
|
NM_004818
|
DDX23
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 23
|
|
chr16_+_56983798
|
1.900
|
NM_001126129
NM_001126130
NM_022770
|
GINS3
|
GINS complex subunit 3 (Psf3 homolog)
|
|
chr19_+_55221023
|
1.897
|
NM_001006656
NM_015428
|
ZNF473
|
zinc finger protein 473
|
|
chr3_+_197951203
|
1.890
|
|
PAK2
|
p21 protein (Cdc42/Rac)-activated kinase 2
|
|
chr2_-_169455167
|
1.887
|
NM_020675
|
SPC25
|
SPC25, NDC80 kinetochore complex component, homolog (S. cerevisiae)
|
|
chr22_+_18447732
|
1.886
|
NM_001190326
NM_022720
|
DGCR8
|
DiGeorge syndrome critical region gene 8
|
|
chr9_-_115212456
|
1.883
|
|
POLE3
|
polymerase (DNA directed), epsilon 3 (p17 subunit)
|
|
chr8_-_145521344
|
1.879
|
NM_012079
|
DGAT1
|
diacylglycerol O-acyltransferase 1
|
|
chr10_+_59815110
|
1.878
|
NM_003201
|
TFAM
|
transcription factor A, mitochondrial
|
|
chr1_+_37930763
|
1.871
|
|
CDCA8
|
cell division cycle associated 8
|
|
chr4_-_2934834
|
1.871
|
|
NOP14
|
NOP14 nucleolar protein homolog (yeast)
|
|
chr17_+_27795633
|
1.862
|
|
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11
|
|
chr8_-_145521297
|
1.861
|
|
DGAT1
|
diacylglycerol O-acyltransferase 1
|
|
chr11_+_61316928
|
1.850
|
|
FEN1
|
flap structure-specific endonuclease 1
|
|
chr1_-_1041598
|
1.836
|
NM_017891
|
C1orf159
|
chromosome 1 open reading frame 159
|
|
chr11_-_64334593
|
1.823
|
NM_130800
NM_130801
|
MEN1
|
multiple endocrine neoplasia I
|
|
chr20_-_32354736
|
1.820
|
NM_000687
|
AHCY
|
adenosylhomocysteinase
|
|
chr11_+_61316989
|
1.808
|
|
FEN1
|
flap structure-specific endonuclease 1
|
|
chr12_-_6831305
|
1.782
|
|
CDCA3
|
cell division cycle associated 3
|
|
chr2_-_74610346
|
1.773
|
|
AUP1
|
ancient ubiquitous protein 1
|
|
chr11_+_61316968
|
1.762
|
|
FEN1
|
flap structure-specific endonuclease 1
|
|
chr11_-_113149642
|
1.752
|
|
ZW10
|
ZW10, kinetochore associated, homolog (Drosophila)
|
|
chr1_-_26105477
|
1.736
|
NM_001145454
NM_005563
|
STMN1
|
stathmin 1
|
|
chr2_-_74610263
|
1.697
|
|
AUP1
|
ancient ubiquitous protein 1
|
|
chr18_-_31178379
|
1.695
|
|
ZNF24
|
zinc finger protein 24
|
|
chr13_-_102224076
|
1.682
|
|
C13orf27
|
chromosome 13 open reading frame 27
|
|
chr9_-_115212393
|
1.681
|
|
POLE3
|
polymerase (DNA directed), epsilon 3 (p17 subunit)
|
|
chr11_+_61316725
|
1.680
|
NM_004111
|
FEN1
|
flap structure-specific endonuclease 1
|
|
chr3_+_32123004
|
1.674
|
NM_015141
|
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like
|
|
chr16_+_23597735
|
1.653
|
|
PLK1
|
polo-like kinase 1
|
|
chr2_-_27288518
|
1.644
|
NM_021095
|
SLC5A6
|
solute carrier family 5 (sodium-dependent vitamin transporter), member 6
|
|
chr3_-_99724438
|
1.643
|
|
CLDND1
|
claudin domain containing 1
|
|
chr16_+_3014028
|
1.642
|
NM_001142350
NM_024339
|
THOC6
|
THO complex 6 homolog (Drosophila)
|
|
chr8_-_145521324
|
1.634
|
|
DGAT1
|
diacylglycerol O-acyltransferase 1
|
|
chr1_+_37930776
|
1.620
|
|
CDCA8
|
cell division cycle associated 8
|
|
chr22_+_28493351
|
1.620
|
NM_001003684
NM_013387
|
UQCR10
|
ubiquinol-cytochrome c reductase, complex III subunit X
|
|
chr16_+_87397686
|
1.608
|
NM_030928
|
CDT1
|
chromatin licensing and DNA replication factor 1
|
|
chr11_-_133628348
|
1.600
|
NM_001037304
NM_001037305
NM_014174
NM_199297
NM_199298
|
THYN1
|
thymocyte nuclear protein 1
|
|
chr18_+_32663056
|
1.578
|
NM_020776
|
KIAA1328
|
KIAA1328
|
|
chr22_+_17847413
|
1.571
|
NM_001178010
NM_001178011
NM_003504
|
CDC45
|
cell division cycle 45 homolog (S. cerevisiae)
|
|
chr12_+_52180971
|
1.570
|
NM_004178
NM_134323
|
TARBP2
|
TAR (HIV-1) RNA binding protein 2
|
|
chr16_+_3014040
|
1.566
|
|
THOC6
|
THO complex 6 homolog (Drosophila)
|
|
chr9_+_115077776
|
1.564
|
NM_004697
|
PRPF4
|
PRP4 pre-mRNA processing factor 4 homolog (yeast)
|
|
chr17_+_7532391
|
1.563
|
NM_001143992
NM_018081
|
WRAP53
|
WD repeat containing, antisense to TP53
|
|
chr16_+_66433882
|
1.560
|
|
THAP11
|
THAP domain containing 11
|
|
chr19_+_3136927
|
1.550
|
|
NCLN
|
nicalin
|
|
chr22_+_20252015
|
1.545
|
|
UBE2L3
|
ubiquitin-conjugating enzyme E2L 3
|
|
chr1_+_155003897
|
1.537
|
NM_005973
|
PRCC
|
papillary renal cell carcinoma (translocation-associated)
|
|
chr16_+_23597661
|
1.536
|
|
PLK1
|
polo-like kinase 1
|
|
chr10_+_5444516
|
1.530
|
NM_001047160
|
NET1
|
neuroepithelial cell transforming 1
|
|
chr5_-_175748145
|
1.526
|
NM_016391
|
NOP16
|
NOP16 nucleolar protein homolog (yeast)
|
|
chr17_+_37694007
|
1.521
|
|
STAT5A
|
signal transducer and activator of transcription 5A
|
|
chr12_-_121551349
|
1.520
|
|
ZCCHC8
|
zinc finger, CCHC domain containing 8
|
|
chr1_-_884483
|
1.518
|
|
NOC2L
|
nucleolar complex associated 2 homolog (S. cerevisiae)
|
|
chr1_+_246087230
|
1.512
|
|
TRIM58
|
tripartite motif containing 58
|
|
chr16_+_23597692
|
1.510
|
NM_005030
|
PLK1
|
polo-like kinase 1
|
|
chr7_+_2361034
|
1.502
|
|
EIF3B
|
eukaryotic translation initiation factor 3, subunit B
|
|
chr17_-_5263706
|
1.489
|
NM_002532
|
NUP88
|
nucleoporin 88kDa
|
|
chr13_-_102224128
|
1.484
|
|
C13orf27
|
chromosome 13 open reading frame 27
|
|
chr9_-_19092648
|
1.483
|
|
HAUS6
|
HAUS augmin-like complex, subunit 6
|
|
chr16_+_23597713
|
1.469
|
|
PLK1
|
polo-like kinase 1
|
|
chr21_+_44351693
|
1.467
|
|
PWP2
|
PWP2 periodic tryptophan protein homolog (yeast)
|
|
chr9_+_115077752
|
1.467
|
|
PRPF4
|
PRP4 pre-mRNA processing factor 4 homolog (yeast)
|
|
chr21_+_44351681
|
1.454
|
|
PWP2
|
PWP2 periodic tryptophan protein homolog (yeast)
|
|
chr5_-_175748533
|
1.448
|
|
NOP16
|
NOP16 nucleolar protein homolog (yeast)
|
|
chr22_+_20251956
|
1.448
|
NM_003347
|
UBE2L3
|
ubiquitin-conjugating enzyme E2L 3
|
|
chr8_-_145612636
|
1.441
|
NM_017767
|
SLC39A4
|
solute carrier family 39 (zinc transporter), member 4
|
|
chr16_+_2807164
|
1.440
|
NM_006799
NM_144956
NM_144957
|
PRSS21
|
protease, serine, 21 (testisin)
|
|
chr1_-_884514
|
1.438
|
NM_015658
|
NOC2L
|
nucleolar complex associated 2 homolog (S. cerevisiae)
|
|
chr7_+_134893185
|
1.431
|
NM_015135
|
NUP205
|
nucleoporin 205kDa
|
|
chr19_+_876984
|
1.426
|
NM_005224
|
ARID3A
|
AT rich interactive domain 3A (BRIGHT-like)
|
|
chr16_+_87397715
|
1.423
|
|
CDT1
|
chromatin licensing and DNA replication factor 1
|
|
chr13_-_102224149
|
1.422
|
NM_138779
|
C13orf27
|
chromosome 13 open reading frame 27
|
|
chr3_-_99724412
|
1.420
|
|
CLDND1
|
claudin domain containing 1
|
|
chr19_+_3136965
|
1.419
|
|
NCLN
|
nicalin
|
|
chr1_+_11963062
|
1.419
|
|
MFN2
|
mitofusin 2
|
|
chr12_-_15005761
|
1.417
|
NM_001175
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta
|
|
chr16_+_3014061
|
1.409
|
|
THOC6
|
THO complex 6 homolog (Drosophila)
|
|
chr6_+_42639929
|
1.404
|
|
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2
|
|
chr16_+_3014052
|
1.399
|
|
THOC6
|
THO complex 6 homolog (Drosophila)
|
|
chr3_-_99724514
|
1.399
|
NM_001040181
NM_001040182
NM_001040183
NM_001040199
NM_001040200
NM_019895
|
CLDND1
|
claudin domain containing 1
|
|
chr22_+_20252026
|
1.395
|
|
UBE2L3
|
ubiquitin-conjugating enzyme E2L 3
|
|
chr12_+_132073014
|
1.394
|
NM_019591
|
ZNF26
|
zinc finger protein 26
|
|
chr6_+_30983941
|
1.394
|
NM_001517
|
GTF2H4
|
general transcription factor IIH, polypeptide 4, 52kDa
|
|
chr8_+_31010319
|
1.393
|
NM_000553
|
WRN
|
Werner syndrome, RecQ helicase-like
|
|
chr3_+_4320036
|
1.389
|
|
SETMAR
|
SET domain and mariner transposase fusion gene
|
|
chr8_+_61753793
|
1.381
|
NM_017780
|
CHD7
|
chromodomain helicase DNA binding protein 7
|
|
chr16_+_23597691
|
1.378
|
|
PLK1
|
polo-like kinase 1
|
|
chr22_+_19078397
|
1.373
|
NM_003426
|
ZNF74
|
zinc finger protein 74
|
|
chr19_-_48976852
|
1.368
|
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4
|
|
chr3_+_4319987
|
1.359
|
NM_006515
|
SETMAR
|
SET domain and mariner transposase fusion gene
|
|
chr2_-_27433130
|
1.347
|
|
GTF3C2
|
general transcription factor IIIC, polypeptide 2, beta 110kDa
|
|
chr11_-_113149602
|
1.346
|
NM_004724
|
ZW10
|
ZW10, kinetochore associated, homolog (Drosophila)
|
|
chr8_-_145521108
|
1.343
|
|
DGAT1
|
diacylglycerol O-acyltransferase 1
|
|
chr1_-_52936602
|
1.338
|
|
C1orf163
|
chromosome 1 open reading frame 163
|
|
chr5_+_85949511
|
1.336
|
|
COX7C
|
cytochrome c oxidase subunit VIIc
|
|
chr3_+_4510030
|
1.334
|
NM_001099952
NM_001168272
NM_002222
|
ITPR1
|
inositol 1,4,5-triphosphate receptor, type 1
|
|
chr19_-_47438518
|
1.329
|
|
GSK3A
|
glycogen synthase kinase 3 alpha
|
|
chr2_-_170138558
|
1.326
|
NM_024622
|
FASTKD1
|
FAST kinase domains 1
|
|
chr17_-_77573974
|
1.326
|
NM_144998
|
STRA13
|
stimulated by retinoic acid 13 homolog (mouse)
|
|
chr15_+_38774676
|
1.323
|
|
RAD51
|
RAD51 homolog (RecA homolog, E. coli) (S. cerevisiae)
|
|
chr19_-_47438548
|
1.323
|
|
GSK3A
|
glycogen synthase kinase 3 alpha
|
|
chr5_-_134397740
|
1.321
|
NM_002653
|
PITX1
|
paired-like homeodomain 1
|
|
chr16_+_517834
|
1.316
|
NM_005632
|
SOLH
|
small optic lobes homolog (Drosophila)
|
|
chr14_+_64523192
|
1.315
|
NM_002028
|
FNTB
|
farnesyltransferase, CAAX box, beta
|
|
chr17_-_45196465
|
1.314
|
|
FAM117A
|
family with sequence similarity 117, member A
|
|
chr1_+_155003939
|
1.313
|
|
PRCC
|
papillary renal cell carcinoma (translocation-associated)
|
|
chr10_-_43390014
|
1.309
|
NM_001099282
NM_001099283
NM_001099284
|
ZNF239
|
zinc finger protein 239
|
|
chr6_-_119712596
|
1.307
|
NM_005907
|
MAN1A1
|
mannosidase, alpha, class 1A, member 1
|
|
chr22_-_27467776
|
1.306
|
NM_001005735
NM_007194
NM_145862
|
CHEK2
|
CHK2 checkpoint homolog (S. pombe)
|
|
chr7_+_2361024
|
1.298
|
|
EIF3B
|
eukaryotic translation initiation factor 3, subunit B
|
|
chr20_-_61600834
|
1.290
|
NM_001958
|
EEF1A2
|
eukaryotic translation elongation factor 1 alpha 2
|
|
chr17_+_27795632
|
1.285
|
|
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11
|
|
chr7_+_149696901
|
1.284
|
|
REPIN1
|
replication initiator 1
|
|
chr4_-_170167962
|
1.282
|
NM_032783
|
CBR4
|
carbonyl reductase 4
|
|
chr6_-_4949237
|
1.278
|
NM_006638
|
RPP40
|
ribonuclease P/MRP 40kDa subunit
|
|
chr11_-_60430430
|
1.272
|
|
PRPF19
|
PRP19/PSO4 pre-mRNA processing factor 19 homolog (S. cerevisiae)
|
|
chr1_+_93317374
|
1.269
|
NM_001164391
NM_001164392
NM_001164393
NM_007358
|
MTF2
|
metal response element binding transcription factor 2
|
|
chr14_+_64523356
|
1.268
|
|
FNTB
|
farnesyltransferase, CAAX box, beta
|
|
chr1_+_45738442
|
1.261
|
NM_015506
|
MMACHC
|
methylmalonic aciduria (cobalamin deficiency) cblC type, with homocystinuria
|
|
chr11_+_61861458
|
1.249
|
|
ASRGL1
|
asparaginase like 1
|
|
chr1_-_52936573
|
1.249
|
|
C1orf163
|
chromosome 1 open reading frame 163
|
|
chr17_+_35697656
|
1.246
|
|
CDC6
|
cell division cycle 6 homolog (S. cerevisiae)
|
|
chr16_-_2037819
|
1.245
|
NM_002528
|
NTHL1
|
nth endonuclease III-like 1 (E. coli)
|