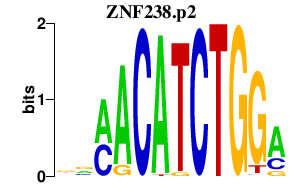
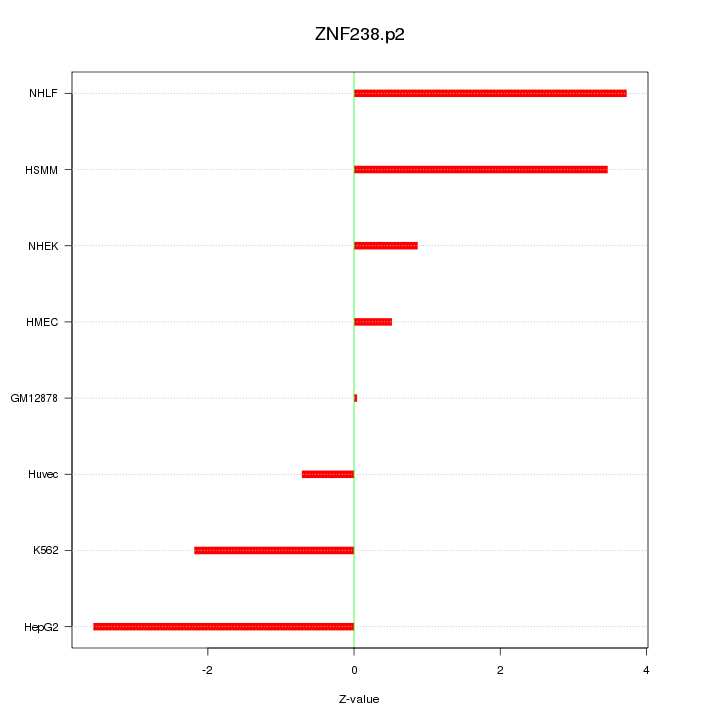
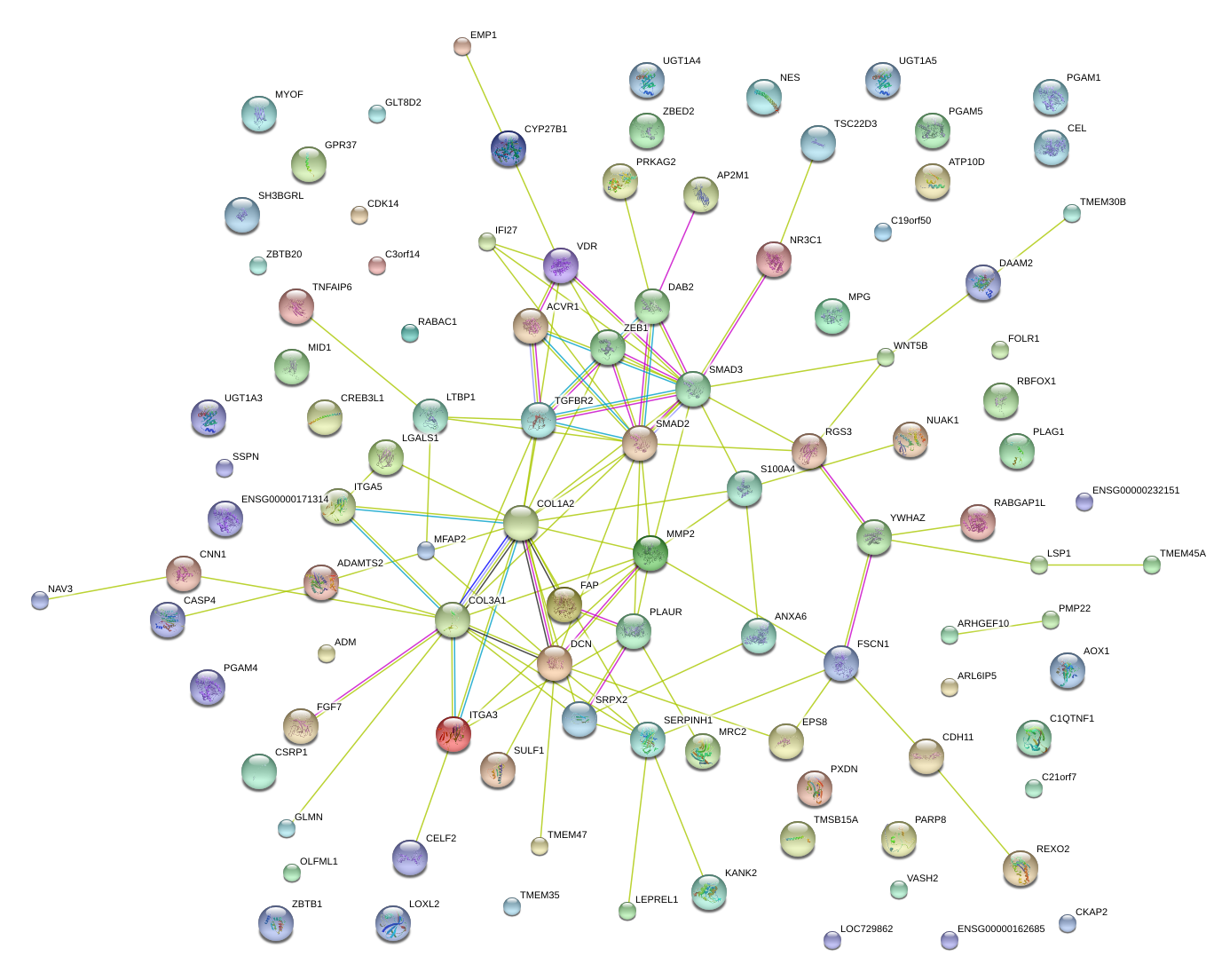
|
chr16_+_54070302
|
10.591
|
NM_004530
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr2_+_189547377
|
7.082
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr2_+_189547323
|
6.976
|
NM_000090
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr2_+_189547358
|
6.959
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr12_+_13240987
|
5.131
|
|
EMP1
|
epithelial membrane protein 1
|
|
chr1_-_151784889
|
4.871
|
NM_002961
NM_019554
|
S100A4
|
S100 calcium binding protein A4
|
|
chr19_-_11166186
|
4.302
|
NM_015493
|
KANK2
|
KN motif and ankyrin repeat domains 2
|
|
chr12_-_105056577
|
4.233
|
|
NUAK1
|
NUAK family, SNF1-like kinase, 1
|
|
chr2_-_162808123
|
4.103
|
NM_004460
|
FAP
|
fibroblast activation protein, alpha
|
|
chr12_-_102968044
|
3.610
|
NM_031302
|
GLT8D2
|
glycosyltransferase 8 domain containing 2
|
|
chr12_-_102967828
|
3.506
|
|
GLT8D2
|
glycosyltransferase 8 domain containing 2
|
|
chr4_+_47181987
|
3.314
|
NM_020453
|
ATP10D
|
ATPase, class V, type 10D
|
|
chrX_-_101658297
|
3.221
|
NM_021992
|
TMSB15A
TMSB15B
|
thymosin beta 15a
thymosin beta 15B
|
|
chr1_-_17179694
|
3.157
|
NM_001135248
NM_002403
|
MFAP2
|
microfibrillar-associated protein 2
|
|
chr9_+_115303527
|
2.809
|
NM_130795
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr1_-_154913712
|
2.632
|
NM_006617
|
NES
|
nestin
|
|
chr3_+_30622905
|
2.578
|
NM_001024847
NM_003242
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa)
|
|
chr12_+_26239755
|
2.543
|
NM_005086
|
SSPN
|
sarcospan (Kras oncogene-associated gene)
|
|
chr10_+_11246934
|
2.420
|
NM_001025076
NM_001083591
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr1_-_146398501
|
2.292
|
|
LOC100286793
FLJ39739
|
hypothetical LOC100286793
hypothetical FLJ39739
|
|
chr10_-_95231933
|
2.264
|
NM_013451
NM_133337
|
MYOF
|
myoferlin
|
|
chr17_+_58058774
|
2.257
|
|
MRC2
|
mannose receptor, C type 2
|
|
chrX_+_99785818
|
2.184
|
NM_014467
|
SRPX2
|
sushi-repeat containing protein, X-linked 2
|
|
chr2_+_151922350
|
2.162
|
NM_007115
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6
|
|
chr1_-_146543102
|
2.145
|
|
|
|
|
chr19_-_48866111
|
2.094
|
NM_001005376
NM_001005377
NM_002659
|
PLAUR
|
plasminogen activator, urokinase receptor
|
|
chrX_+_99786044
|
2.081
|
|
SRPX2
|
sushi-repeat containing protein, X-linked 2
|
|
chr1_-_143232526
|
2.081
|
|
FAM91A2
|
family with sequence similarity 91, member A2
|
|
chr12_+_76749199
|
2.060
|
NM_014903
|
NAV3
|
neuron navigator 3
|
|
chr6_+_39868770
|
2.034
|
NM_015345
|
DAAM2
|
dishevelled associated activator of morphogenesis 2
|
|
chr17_+_45488330
|
2.009
|
NM_002204
NM_005501
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor)
|
|
chr1_-_142535878
|
2.004
|
|
LOC100286793
FLJ39739
|
hypothetical LOC100286793
hypothetical FLJ39739
|
|
chr3_+_101694098
|
1.954
|
NM_018004
|
TMEM45A
|
transmembrane protein 45A
|
|
chr1_-_146543016
|
1.951
|
|
|
|
|
chr12_+_26239535
|
1.951
|
NM_001135823
|
SSPN
|
sarcospan (Kras oncogene-associated gene)
|
|
chr2_+_201182187
|
1.933
|
|
AOX1
|
aldehyde oxidase 1
|
|
chr3_-_191321601
|
1.883
|
NM_018192
|
LEPREL1
|
leprecan-like 1
|
|
chr3_+_101694244
|
1.879
|
|
TMEM45A
|
transmembrane protein 45A
|
|
chr1_-_142536082
|
1.862
|
|
|
|
|
chr17_+_45488356
|
1.847
|
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor)
|
|
chr8_+_1759502
|
1.832
|
NM_014629
|
ARHGEF10
|
Rho guanine nucleotide exchange factor (GEF) 10
|
|
chrX_-_106905545
|
1.687
|
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr7_+_90063543
|
1.663
|
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr12_-_15833383
|
1.519
|
|
EPS8
|
epidermal growth factor receptor pathway substrate 8
|
|
chr6_+_39868126
|
1.492
|
|
DAAM2
|
dishevelled associated activator of morphogenesis 2
|
|
chr21_+_29439768
|
1.492
|
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chr5_+_49998677
|
1.482
|
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8
|
|
chr5_+_49998528
|
1.458
|
NM_001178056
NM_024615
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8
|
|
chr2_-_1727324
|
1.453
|
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chrX_+_100220491
|
1.364
|
NM_021637
|
TMEM35
|
transmembrane protein 35
|
|
chr17_-_15109315
|
1.348
|
NM_000304
|
PMP22
|
peripheral myelin protein 22
|
|
chr1_-_199742871
|
1.293
|
NM_001193570
NM_004078
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chrX_-_106905657
|
1.275
|
NM_198057
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chrX_+_80344253
|
1.266
|
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like
|
|
chr2_-_1727247
|
1.255
|
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr1_+_211190476
|
1.252
|
NM_001136474
NM_001136475
NM_024749
|
VASH2
|
vasohibin 2
|
|
chr1_+_154291140
|
1.234
|
NM_001145264
NM_014017
|
LAMTOR2
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2
|
|
chr16_-_63713466
|
1.230
|
|
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast)
|
|
chr14_+_64041257
|
1.227
|
|
ZBTB1
|
zinc finger and BTB domain containing 1
|
|
chr11_+_7463175
|
1.204
|
NM_198474
|
OLFML1
|
olfactomedin-like 1
|
|
chr3_+_30623376
|
1.204
|
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa)
|
|
chr22_+_36401558
|
1.203
|
NM_002305
|
LGALS1
|
lectin, galactoside-binding, soluble, 1
|
|
chr3_+_62280435
|
1.198
|
NM_020685
|
C3orf14
|
chromosome 3 open reading frame 14
|
|
chr2_-_158439868
|
1.188
|
NM_001105
|
ACVR1
|
activin A receptor, type I
|
|
chr2_-_1727292
|
1.167
|
NM_012293
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr8_+_70541412
|
1.165
|
NM_001128204
NM_015170
|
SULF1
|
sulfatase 1
|
|
chr5_-_178704934
|
1.165
|
|
ADAMTS2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2
|
|
chr7_-_124192916
|
1.163
|
NM_005302
|
GPR37
|
G protein-coupled receptor 37 (endothelin receptor type B-like)
|
|
chr11_+_74950766
|
1.153
|
NM_001235
|
SERPINH1
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1)
|
|
chr11_+_10283171
|
1.143
|
NM_001124
|
ADM
|
adrenomedullin
|
|
chrX_-_34585287
|
1.112
|
NM_031442
|
TMEM47
|
transmembrane protein 47
|
|
chr1_-_199742864
|
1.104
|
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chr16_-_63713485
|
1.084
|
|
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast)
|
|
chr11_-_104332631
|
1.054
|
NM_033306
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase
|
|
chr19_-_47155340
|
1.043
|
NM_006423
|
RABAC1
|
Rab acceptor 1 (prenylated)
|
|
chr12_-_15833617
|
1.026
|
NM_004447
|
EPS8
|
epidermal growth factor receptor pathway substrate 8
|
|
chr3_+_69216747
|
1.017
|
NM_006407
|
ARL6IP5
|
ADP-ribosylation-like factor 6 interacting protein 5
|
|
chr14_+_93646831
|
1.013
|
NM_001130080
NM_005532
|
IFI27
|
interferon, alpha-inducible protein 27
|
|
chr17_+_74531820
|
1.009
|
NM_030968
NM_198594
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1
|
|
chr7_-_151142858
|
0.981
|
NM_001040633
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit
|
|
chr1_-_143232344
|
0.979
|
|
LOC728855
|
hypothetical LOC728855
|
|
chr1_-_199742841
|
0.976
|
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chr12_-_46585032
|
0.951
|
NM_000376
NM_001017535
|
VDR
|
vitamin D (1,25- dihydroxyvitamin D3) receptor
|
|
chr17_+_58058783
|
0.951
|
|
MRC2
|
mannose receptor, C type 2
|
|
chr11_+_113815317
|
0.939
|
NM_015523
|
REXO2
|
REX2, RNA exonuclease 2 homolog (S. cerevisiae)
|
|
chr5_-_178705036
|
0.933
|
NM_014244
NM_021599
|
ADAMTS2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2
|
|
chr1_-_143232464
|
0.928
|
|
|
|
|
chr9_+_115396020
|
0.907
|
NM_144489
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr1_-_143232275
|
0.900
|
|
LOC728855
|
hypothetical LOC728855
|
|
chr5_-_142764168
|
0.896
|
NM_001018076
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|
|
chr16_+_6009095
|
0.886
|
NM_001142333
NM_018723
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr11_+_74950893
|
0.882
|
|
SERPINH1
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1)
|
|
chr8_-_23233426
|
0.879
|
|
LOXL2
|
lysyl oxidase-like 2
|
|
chr5_-_150501393
|
0.874
|
NM_001193544
|
ANXA6
|
annexin A6
|
|
chr8_-_102029997
|
0.868
|
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
|
|
chr7_+_93862137
|
0.861
|
|
COL1A2
PLAG1
|
collagen, type I, alpha 2
pleiomorphic adenoma gene 1
|
|
chr3_+_185377341
|
0.822
|
|
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit
|
|
chr19_+_11510578
|
0.798
|
NM_001299
|
CNN1
|
calponin 1, basic, smooth muscle
|
|
chr11_+_46256130
|
0.797
|
|
CREB3L1
|
cAMP responsive element binding protein 3-like 1
|
|
chr14_-_60818263
|
0.784
|
NM_001017970
|
TMEM30B
|
transmembrane protein 30B
|
|
chr19_+_18529652
|
0.776
|
|
C19orf50
|
chromosome 19 open reading frame 50
|
|
chrX_-_10548458
|
0.734
|
NM_000381
NM_033289
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr12_-_53099269
|
0.730
|
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr19_+_18529571
|
0.719
|
NM_001171948
NM_001171949
NM_024069
|
C19orf50
|
chromosome 19 open reading frame 50
|
|
chr2_+_33213111
|
0.718
|
NM_000627
NM_001166264
NM_001166265
NM_001166266
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chr3_-_115585178
|
0.717
|
NM_001164342
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr12_-_90097384
|
0.715
|
NM_133503
|
DCN
|
decorin
|
|
chr2_+_234302511
|
0.711
|
NM_019093
|
UGT1A3
|
UDP glucuronosyltransferase 1 family, polypeptide A3
|
|
chr3_+_185377430
|
0.710
|
|
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit
|
|
chr13_+_51933228
|
0.707
|
|
CKAP2
|
cytoskeleton associated protein 2
|
|
chr16_-_63713382
|
0.703
|
NM_001797
|
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast)
|
|
chr5_-_39460787
|
0.689
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr15_+_47502666
|
0.667
|
NM_002009
|
FGF7
|
fibroblast growth factor 7
|
|
chr12_+_1608672
|
0.665
|
NM_032642
|
WNT5B
|
wingless-type MMTV integration site family, member 5B
|
|
chr7_+_5609416
|
0.664
|
|
FSCN1
|
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus)
|
|
chr11_+_71578249
|
0.633
|
NM_016724
NM_016725
|
FOLR1
|
folate receptor 1 (adult)
|
|
chr14_+_64040979
|
0.623
|
NM_001123329
NM_014950
|
ZBTB1
|
zinc finger and BTB domain containing 1
|
|
chr10_+_31647425
|
0.613
|
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr19_-_47155277
|
0.609
|
|
RABAC1
|
Rab acceptor 1 (prenylated)
|
|
chr11_+_1846478
|
0.603
|
NM_001013253
|
LSP1
|
lymphocyte-specific protein 1
|
|
chrX_-_77111790
|
0.599
|
NM_001029891
|
PGAM4
|
phosphoglycerate mutase family member 4
|
|
chr17_+_74531873
|
0.573
|
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1
|
|
chr1_+_173111306
|
0.572
|
NM_001035230
|
RABGAP1L
|
RAB GTPase activating protein 1-like
|
|
chr3_-_112796855
|
0.570
|
NM_024508
|
ZBED2
|
zinc finger, BED-type containing 2
|
|
chr15_+_65205107
|
0.557
|
NM_001145102
|
SMAD3
|
SMAD family member 3
|
|
chr5_-_83052606
|
0.550
|
NM_001884
|
HAPLN1
|
hyaluronan and proteoglycan link protein 1
|
|
chr11_+_46255737
|
0.547
|
NM_052854
|
CREB3L1
|
cAMP responsive element binding protein 3-like 1
|
|
chr3_-_191321367
|
0.545
|
|
LEPREL1
|
leprecan-like 1
|
|
chr14_-_19090251
|
0.534
|
NM_001145442
|
POTEG
POTEM
|
POTE ankyrin domain family, member G
POTE ankyrin domain family, member M
|
|
chr19_+_55627971
|
0.523
|
NM_004533
|
MYBPC2
|
myosin binding protein C, fast type
|
|
chr7_+_154493478
|
0.523
|
NM_024012
|
HTR5A
|
5-hydroxytryptamine (serotonin) receptor 5A
|
|
chr4_+_114190233
|
0.515
|
NM_001148
NM_020977
|
ANK2
|
ankyrin 2, neuronal
|
|
chr5_-_39460687
|
0.511
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr13_-_32678142
|
0.510
|
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr5_+_40715778
|
0.507
|
NM_000958
|
PTGER4
|
prostaglandin E receptor 4 (subtype EP4)
|
|
chr12_-_53099203
|
0.502
|
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr22_-_35011703
|
0.495
|
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr12_-_53099277
|
0.490
|
NM_002205
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr5_-_151046638
|
0.488
|
|
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin)
|
|
chr3_+_49034494
|
0.486
|
|
NDUFAF3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 3
|
|
chr21_-_38210531
|
0.484
|
NM_002240
|
KCNJ6
|
potassium inwardly-rectifying channel, subfamily J, member 6
|
|
chr3_-_52248071
|
0.480
|
|
TWF2
|
twinfilin, actin-binding protein, homolog 2 (Drosophila)
|
|
chr5_-_41906459
|
0.479
|
NM_000436
|
OXCT1
|
3-oxoacid CoA transferase 1
|
|
chr11_+_5574440
|
0.474
|
NM_001003818
NM_001198644
NM_001198645
NM_001003819
|
TRIM6
TRIM6-TRIM34
|
tripartite motif containing 6
TRIM6-TRIM34 readthrough
|
|
chr17_-_51164443
|
0.456
|
NM_001099640
|
TMEM100
|
transmembrane protein 100
|
|
chr19_-_50518072
|
0.456
|
NM_001824
|
CKM
|
creatine kinase, muscle
|
|
chr11_+_32068990
|
0.451
|
NM_002901
|
RCN1
|
reticulocalbin 1, EF-hand calcium binding domain
|
|
chr1_+_181422016
|
0.451
|
|
LAMC2
|
laminin, gamma 2
|
|
chr1_-_199742589
|
0.447
|
NM_001193571
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chr4_+_157044296
|
0.434
|
NM_005651
|
TDO2
|
tryptophan 2,3-dioxygenase
|
|
chr5_+_140762551
|
0.434
|
NM_018921
NM_032089
|
PCDHGA9
|
protocadherin gamma subfamily A, 9
|
|
chr17_+_7732759
|
0.428
|
NM_001005273
NM_005852
|
CHD3
|
chromodomain helicase DNA binding protein 3
|
|
chr11_-_57092332
|
0.425
|
|
UBE2L6
|
ubiquitin-conjugating enzyme E2L 6
|
|
chr1_-_238842071
|
0.422
|
NM_022469
|
GREM2
|
gremlin 2
|
|
chr12_+_111047676
|
0.419
|
NM_001143906
NM_006700
|
TRAFD1
|
TRAF-type zinc finger domain containing 1
|
|
chr21_+_46226072
|
0.418
|
NM_001848
|
COL6A1
|
collagen, type VI, alpha 1
|
|
chr20_+_17498507
|
0.405
|
NM_001011546
NM_006870
|
DSTN
|
destrin (actin depolymerizing factor)
|
|
chr1_+_53165576
|
0.392
|
|
SCP2
|
sterol carrier protein 2
|
|
chr7_+_100113664
|
0.370
|
|
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2
|
|
chr2_+_131692393
|
0.370
|
NM_001083538
|
POTEE
|
POTE ankyrin domain family, member E
|
|
chr3_-_52248182
|
0.369
|
NM_007284
|
TWF2
|
twinfilin, actin-binding protein, homolog 2 (Drosophila)
|
|
chr12_-_54408391
|
0.369
|
|
CD63
|
CD63 molecule
|
|
chr12_+_111047742
|
0.368
|
|
TRAFD1
|
TRAF-type zinc finger domain containing 1
|
|
chr15_+_53398424
|
0.368
|
NM_004855
|
PIGB
|
phosphatidylinositol glycan anchor biosynthesis, class B
|
|
chr13_-_29322159
|
0.367
|
|
UBL3
|
ubiquitin-like 3
|
|
chr17_-_64649608
|
0.367
|
NM_080284
|
ABCA6
|
ATP-binding cassette, sub-family A (ABC1), member 6
|
|
chr1_-_201586872
|
0.362
|
|
|
|
|
chr5_+_140772691
|
0.358
|
NM_018913
NM_032090
|
PCDHGA10
|
protocadherin gamma subfamily A, 10
|
|
chr15_+_60726801
|
0.356
|
NM_015059
|
TLN2
|
talin 2
|
|
chr18_+_7557313
|
0.356
|
NM_001105244
NM_002845
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M
|
|
chr2_-_16710551
|
0.350
|
NM_030797
|
FAM49A
|
family with sequence similarity 49, member A
|
|
chr14_+_95792299
|
0.347
|
NM_000710
|
BDKRB1
|
bradykinin receptor B1
|
|
chr5_+_162862808
|
0.344
|
NM_182796
|
MAT2B
|
methionine adenosyltransferase II, beta
|
|
chr2_-_235070431
|
0.328
|
NM_005737
|
ARL4C
|
ADP-ribosylation factor-like 4C
|
|
chr3_+_49034050
|
0.328
|
NM_199069
NM_199070
|
NDUFAF3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 3
|
|
chr15_+_29406335
|
0.324
|
NM_015995
|
KLF13
|
Kruppel-like factor 13
|
|
chr14_+_77939845
|
0.322
|
NM_004796
|
NRXN3
|
neurexin 3
|
|
chr12_-_107257205
|
0.314
|
NM_001142343
NM_001142344
NM_004072
|
CMKLR1
|
chemokine-like receptor 1
|
|
chr11_+_32069183
|
0.313
|
|
RCN1
|
reticulocalbin 1, EF-hand calcium binding domain
|
|
chr4_+_154345039
|
0.310
|
NM_015271
|
TRIM2
|
tripartite motif containing 2
|
|
chr10_+_116843108
|
0.309
|
NM_207303
|
ATRNL1
|
attractin-like 1
|
|
chr10_+_6284845
|
0.298
|
NM_004566
|
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3
|
|
chr6_-_36623223
|
0.296
|
NM_007271
|
STK38
|
serine/threonine kinase 38
|
|
chr10_-_27189898
|
0.294
|
|
ABI1
|
abl-interactor 1
|
|
chr11_+_111288686
|
0.291
|
|
HSPB2-C11ORF52
HSPB2
|
HSPB2-C11orf52 read-through transcript
heat shock 27kDa protein 2
|
|
chr8_+_79590890
|
0.286
|
NM_006823
NM_181839
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha
|
|
chr6_+_163068991
|
0.284
|
NM_001080379
|
PACRG
|
PARK2 co-regulated
|
|
chr10_-_27189797
|
0.282
|
|
ABI1
|
abl-interactor 1
|
|
chr2_+_187173176
|
0.277
|
NM_001144999
|
ITGAV
|
integrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51)
|
|
chr1_-_85816520
|
0.277
|
NM_001134445
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr13_+_96672705
|
0.272
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr10_-_27189995
|
0.270
|
NM_001012750
NM_001012751
NM_001012752
NM_001178116
NM_001178119
NM_001178120
NM_001178121
NM_001178122
NM_001178123
NM_001178124
NM_001178125
NM_005470
|
ABI1
|
abl-interactor 1
|
|
chr3_-_15814678
|
0.269
|
NM_001195098
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr5_+_1854493
|
0.265
|
NM_004553
|
NDUFS6
|
NADH dehydrogenase (ubiquinone) Fe-S protein 6, 13kDa (NADH-coenzyme Q reductase)
|
|
chr10_-_69422391
|
0.265
|
|
HERC4
|
hect domain and RLD 4
|
|
chr1_+_144287368
|
0.252
|
|
PIAS3
|
protein inhibitor of activated STAT, 3
|
|
chr19_+_8384181
|
0.248
|
NM_001005415
NM_001005416
NM_016496
|
MARCH2
|
membrane-associated ring finger (C3HC4) 2
|
|
chr14_+_89934358
|
0.245
|
NM_001166106
|
CALM1
|
calmodulin 1 (phosphorylase kinase, delta)
|
|
chr7_+_141342147
|
0.245
|
NM_004668
|
MGAM
|
maltase-glucoamylase (alpha-glucosidase)
|
|
chr20_-_6051562
|
0.244
|
|
FERMT1
|
fermitin family member 1
|