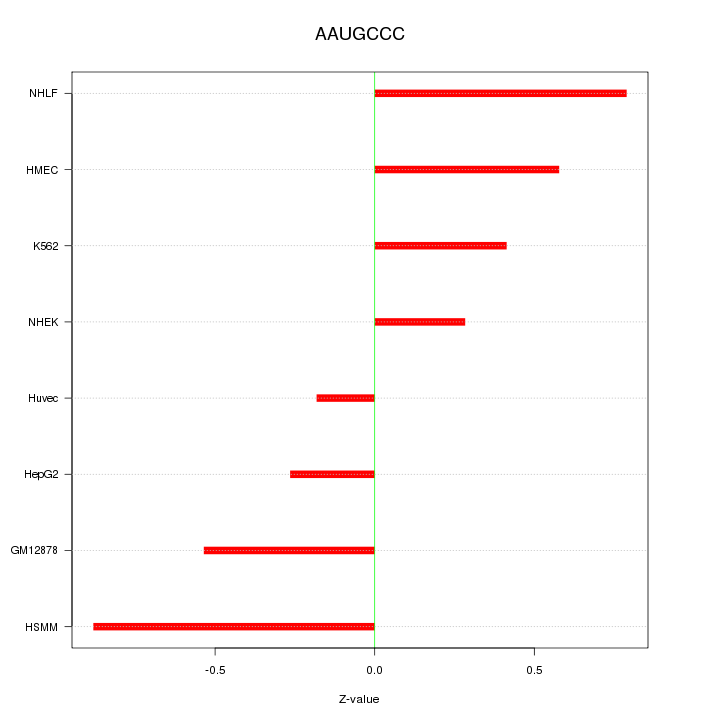
| 0.1 |
0.3 |
GO:0043371 |
negative regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043371) negative regulation of T-helper cell differentiation(GO:0045623) negative regulation of CD4-positive, alpha-beta T cell activation(GO:2000515) |
| 0.1 |
0.4 |
GO:0060414 |
aorta development(GO:0035904) aorta morphogenesis(GO:0035909) aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 |
0.2 |
GO:0009642 |
response to light intensity(GO:0009642) response to vitamin E(GO:0033197) |
| 0.0 |
0.2 |
GO:0060839 |
radial pattern formation(GO:0009956) lymphatic endothelial cell fate commitment(GO:0060838) endothelial cell fate commitment(GO:0060839) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.0 |
0.2 |
GO:0070208 |
protein heterotrimerization(GO:0070208) |
| 0.0 |
0.3 |
GO:0060666 |
dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 |
0.3 |
GO:0060347 |
heart trabecula formation(GO:0060347) heart trabecula morphogenesis(GO:0061384) |
| 0.0 |
0.2 |
GO:0090025 |
regulation of monocyte chemotaxis(GO:0090025) positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 |
0.1 |
GO:0050774 |
negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 |
0.2 |
GO:0043982 |
histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 |
0.1 |
GO:0046984 |
regulation of hemoglobin biosynthetic process(GO:0046984) |
| 0.0 |
0.1 |
GO:0043402 |
glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 |
0.3 |
GO:0060638 |
mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.0 |
0.1 |
GO:0071460 |
cellular response to cell-matrix adhesion(GO:0071460) |
| 0.0 |
0.0 |
GO:0060061 |
Spemann organizer formation(GO:0060061) |
| 0.0 |
0.0 |
GO:0043983 |
histone H4-K12 acetylation(GO:0043983) |
| 0.0 |
0.0 |
GO:0060017 |
parathyroid gland development(GO:0060017) |
| 0.0 |
0.1 |
GO:0035283 |
rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 |
0.1 |
GO:0048625 |
myoblast fate commitment(GO:0048625) |