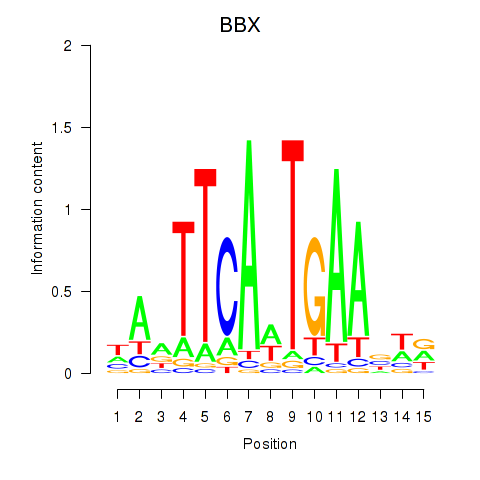
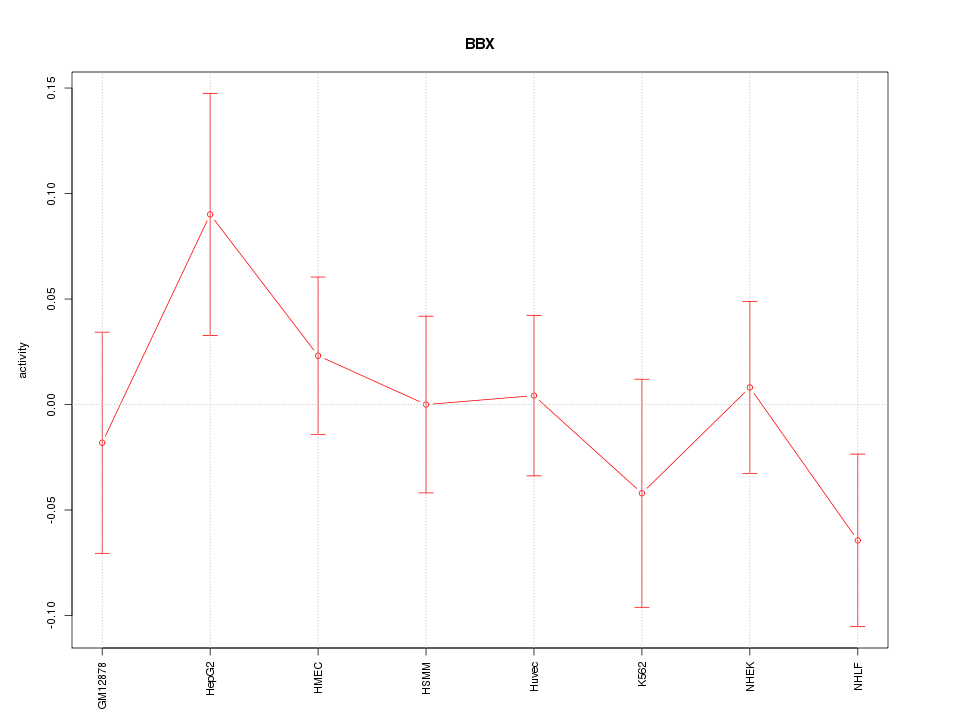
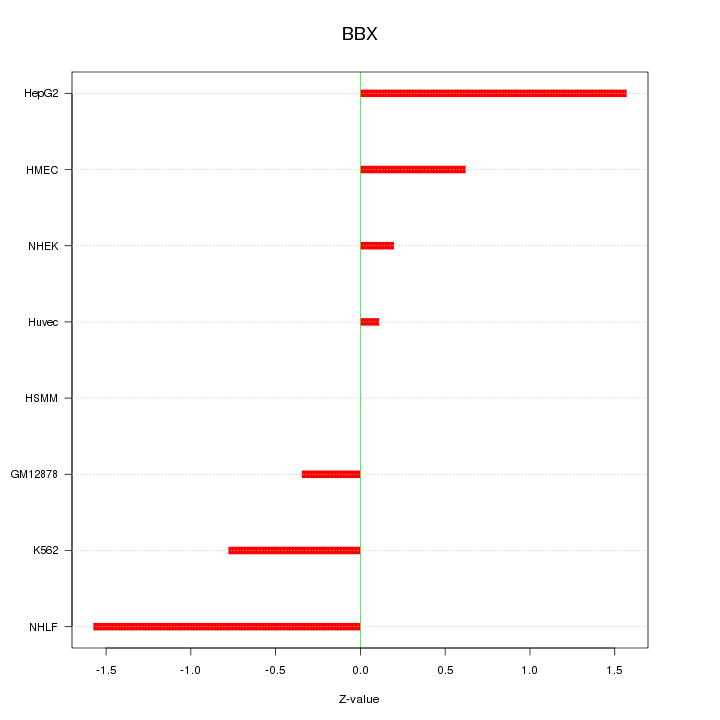
| 0.2 |
2.1 |
GO:0006787 |
porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 |
0.4 |
GO:0043461 |
proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.1 |
0.3 |
GO:0007206 |
phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.1 |
0.6 |
GO:0046898 |
response to cycloheximide(GO:0046898) |
| 0.1 |
0.3 |
GO:0045475 |
locomotor rhythm(GO:0045475) |
| 0.1 |
1.5 |
GO:0008210 |
estrogen metabolic process(GO:0008210) |
| 0.0 |
4.1 |
GO:0051592 |
response to calcium ion(GO:0051592) |
| 0.0 |
0.2 |
GO:0015712 |
hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 |
0.2 |
GO:0070254 |
mucus secretion(GO:0070254) |
| 0.0 |
0.2 |
GO:0071108 |
protein K48-linked deubiquitination(GO:0071108) |
| 0.0 |
0.7 |
GO:0007398 |
ectoderm development(GO:0007398) |
| 0.0 |
0.6 |
GO:0036503 |
ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) ERAD pathway(GO:0036503) protein autoubiquitination(GO:0051865) |
| 0.0 |
0.1 |
GO:0034287 |
detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 |
0.9 |
GO:0006958 |
complement activation, classical pathway(GO:0006958) |
| 0.0 |
0.6 |
GO:0019433 |
triglyceride catabolic process(GO:0019433) |
| 0.0 |
0.1 |
GO:2000172 |
regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.0 |
0.1 |
GO:0007064 |
mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 |
0.4 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 |
0.1 |
GO:0035964 |
COPI-coated vesicle budding(GO:0035964) Golgi vesicle budding(GO:0048194) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 |
0.0 |
GO:0046532 |
regulation of photoreceptor cell differentiation(GO:0046532) |